Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Class Definition: KDTreeSearcher
statistics: KDTreeSearcher
KD-tree nearest neighbor searcher
The KDTreeSearcher class implements a KD-tree search algorithm for
nearest neighbor queries. It stores training data and supports various
distance metrics along with their parameter values for performing a KD-tree
search. The KD-tree algorithm partitions the training data into a
hierarchical tree structure and performs search operations by traversing
the tree to reduce the number of distance computations. It facilitates
nearest neighbor queries using knnsearch and radius queries using
rangesearch.
You can either use the KDTreeSearcher class constructor or the
createns function to create an KDTreeSearcher object.
See also: createns, ExhaustiveSearcher, hnswSearcher, knnsearch, rangesearch
Source Code: KDTreeSearcher
Properties
Point data, specified as an numeric matrix where each row is an observation and each column is a feature. This property is private and cannot be modified after object creation.
The maximum number of data points in the leaf node of the KD-tree. Default value is 50. This property is private and cannot be modified after object creation.
Distance metric used for searches, specified as a character vector.
Supported metrics are "euclidean", "cityblock",
"minkowski", and "chebychev". Default value is
"euclidean".
The type and value of the distance parameter depends on the selected
Distance metric and can be any of the following:
- For
"minkowski", a positive scalar exponent (default 2). - Empty for other metrics (
"euclidean","cityblock","chebychev"). Attempting to set a non-empty value for these metrics will result in an error.
Methods
KDTreeSearcher: obj = KDTreeSearcher (X)
KDTreeSearcher: obj = KDTreeSearcher (X, name, value)
obj = KDTreeSearcher (X) constructs a
KDTreeSearcher object with training data X using the
default "euclidean" distance metric. X must be an
numeric matrix, where rows represent observations and columns
represent features.
obj = KDTreeSearcher (X, name, value)
allows customization through name-value pairs:
| Name | Value | |
|---|---|---|
"Distance" | Distance metric, specified as a
character vector ("euclidean", "cityblock",
"minkowski", "chebychev"). Default is
"euclidean". | |
"P" | Minkowski distance exponent, a positive
scalar. Valid only when "Distance" is "minkowski".
Default is 2. | |
"BucketSize" | Maximum number of data points in the leaf node of the KD-tree, a positive integer. Default is 50. |
You can also create a KDTreeSearcher object using the
createns function.
See also: KDTreeSearcher, knnsearch, rangesearch, createns
KDTreeSearcher: [idx, D] = knnsearch (obj, Y)
KDTreeSearcher: [idx, D] = knnsearch (obj, Y, name, value)
[idx, D] = knnsearch (obj, Y, K)
returns the indices idx and distances D of the
nearest neighbors in obj.X to each point in Y, using the
distance metric specified in obj.Distance.
- obj is a
KDTreeSearcherobject. - Y is an numeric matrix of query points, where must match the number of columns in obj.X.
- idx contains the indices of the nearest neighbors in obj.X.
- D contains the corresponding distances.
[idx, D] = knnsearch (obj, Y, name,
value) allows additional options via name-value pairs:
| Name | Value | |
|---|---|---|
"K" | A positive integer specifying the number of nearest neighbors to find. Default is 1. | |
"IncludeTies" | Logical flag indicating whether to
include all neighbors tied with the th smallest distance. Default
is false. If true, idx and D are cell arrays. | |
"SortIndices" | Logical flag indicating whether to
sort the indices by distance. Default is true. |
See also: KDTreeSearcher, rangesearch
KDTreeSearcher: [idx, D] = rangesearch (obj, Y, r)
KDTreeSearcher: [idx, D] = rangesearch (obj, Y, r, name, value)
[idx, D] = rangesearch (obj, Y, r)
returns the indices idx and distances D of all points in
obj.X within radius r of each point in Y, using the
distance metric specified in obj.Distance.
- obj is a
KDTreeSearcherobject. - Y is an numeric matrix of query points, where must match the number of columns in obj.X.
- r is a nonnegative scalar specifying the search radius.
[idx, D] = rangesearch (obj, Y, r, name, value)
allows additional options via name-value pairs:
| Name | Value | |
|---|---|---|
"SortIndices" | Logical flag indicating whether to
sort the indices by distance. Default is true. |
idx and D are cell arrays where each cell contains the indices and distances for one query point in Y.
See also: KDTreeSearcher, knnsearch
Examples
load fisheriris numSamples = size (meas, 1); queryIndices = [1, 23, 46, 63, 109]; dataIndices = ~ismember (1:numSamples, queryIndices); queryPoints = meas(queryIndices, :); dataPoints = meas(dataIndices, :); searchRadius = 0.3; kdTree = KDTreeSearcher (dataPoints, 'Distance', 'minkowski') nearestNeighbors = knnsearch (kdTree, queryPoints, "K", 2) neighborsInRange = rangesearch (kdTree, queryPoints, searchRadius) |
kdTree =
KDTreeSearcher with properties:
BucketSize: 50
Distance: 'minkowski'
DistParameter: 2
X: [145x4 double]
nearestNeighbors =
17 4
6 2
1 12
89 66
124 100
neighborsInRange =
5x1 cell array
{9x1 double}
{0x1 double}
{7x1 double}
{0x1 double}
{0x1 double}
|
X = [1, 2; 3, 4; 5, 6];
obj = KDTreeSearcher (X);
## Find the nearest neighbor to [2, 3]
Y = [2, 3];
[idx, D] = knnsearch (obj, Y, "K", 1);
disp ("Nearest neighbor index:");
disp (idx);
disp ("Distance:");
disp (D);
## Find all points within radius 2
[idx, D] = rangesearch (obj, Y, 2);
disp ("Indices within radius:");
disp (idx);
disp ("Distances:");
disp (D); |
Nearest neighbor index:
1
Distance:
1.4142
Indices within radius:
1x1 cell array
{2x1 double}
Distances:
1x1 cell array
{2x1 double}
|
X = [0, 0; 1, 0; 2, 0];
obj = KDTreeSearcher (X, "Distance", "minkowski", "P", 3);
## Find the nearest neighbor to [1, 0]
Y = [1, 0];
[idx, D] = knnsearch (obj, Y, "K", 1);
disp ("Nearest neighbor index:");
disp (idx);
disp ("Distance:");
disp (D); |
Nearest neighbor index: 2 Distance: 0 |
rng(42);
disp('Demonstrating KDTreeSearcher');
n = 100;
mu1 = [0.3, 0.3];
mu2 = [0.7, 0.7];
sigma = 0.1;
X1 = mu1 + sigma * randn (n / 2, 2);
X2 = mu2 + sigma * randn (n / 2, 2);
X = [X1; X2];
obj = KDTreeSearcher(X);
Y = [0.3, 0.3; 0.7, 0.7; 0.5, 0.5];
K = 5;
[idx, D] = knnsearch (obj, Y, "K", K);
disp ('For the first query point:');
disp (['Query point: ', num2str(Y(1,:))]);
disp ('Indices of nearest neighbors:');
disp (idx(1,:));
disp ('Distances:');
disp (D(1,:));
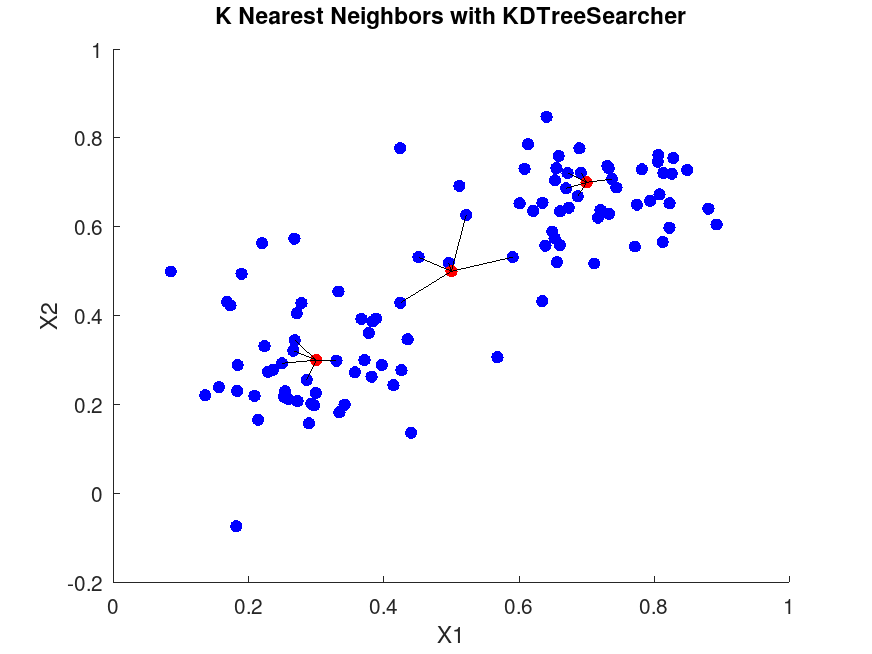
figure;
scatter (X(:,1), X(:,2), 36, 'b', 'filled'); # Training points
hold on;
scatter (Y(:,1), Y(:,2), 36, 'r', 'filled'); # Query points
for i = 1:size (Y, 1)
query = Y(i,:);
neighbors = X(idx(i,:), :);
for j = 1:K
plot ([query(1), neighbors(j,1)], [query(2), neighbors(j,2)], 'k-');
endfor
endfor
hold off;
title ('K Nearest Neighbors with KDTreeSearcher');
xlabel ('X1');
ylabel ('X2');
r = 0.15;
[idx, D] = rangesearch (obj, Y, r);
disp ('For the first query point in rangesearch:');
disp (['Query point: ', num2str(Y(1,:))]);
disp ('Indices of points within radius:');
disp (idx{1});
disp ('Distances:');
disp (D{1});
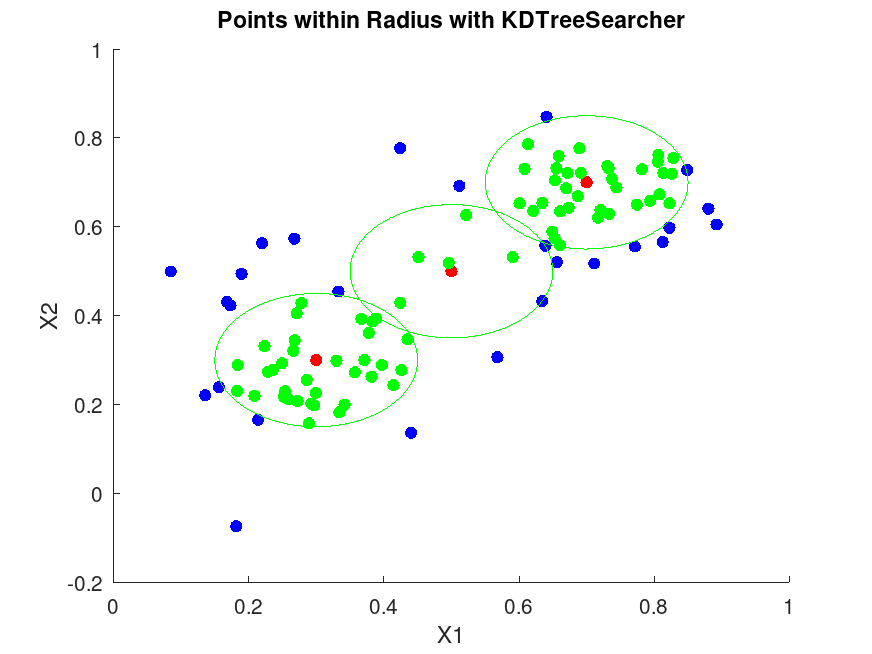
figure;
scatter (X(:,1), X(:,2), 36, 'b', 'filled');
hold on;
scatter (Y(:,1), Y(:,2), 36, 'r', 'filled');
theta = linspace (0, 2 * pi, 100);
for i = 1:size (Y, 1)
center = Y(i,:);
x_circle = center(1) + r * cos (theta);
y_circle = center(2) + r * sin (theta);
plot (x_circle, y_circle, 'g-');
## Highlight points within radius
if (! isempty (idx{i}))
in_radius = X(idx{i}, :);
scatter (in_radius(:,1), in_radius(:,2), 36, 'g', 'filled');
endif
endfor
hold off
title ('Points within Radius with KDTreeSearcher');
xlabel ('X1');
ylabel ('X2'); |
Demonstrating KDTreeSearcher
For the first query point:
Query point: 0.3 0.3
Indices of nearest neighbors:
49 19 14 46 34
Distances:
0.029932 0.040026 0.046845 0.051107 0.054789
For the first query point in rangesearch:
Query point: 0.3 0.3
Indices of points within radius:
49
19
14
46
34
23
12
41
1
3
42
48
24
2
11
37
10
27
32
20
44
40
39
21
7
31
45
30
4
16
47
9
29
8
Distances:
0.029932
0.040026
0.046845
0.051107
0.054789
0.063517
0.067855
0.071365
0.073769
0.075991
0.082686
0.084066
0.090008
0.095171
0.096337
0.096836
0.097593
0.098255
0.098948
0.101780
0.108652
0.108983
0.114272
0.116760
0.120198
0.121721
0.122560
0.127342
0.128062
0.128687
0.130208
0.136007
0.142870
0.143009 |