Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: fitcgam
statistics: Mdl = fitcgam (X, Y)
statistics: Mdl = fitcgam (…, name, value)
Fit a Generalized Additive Model (GAM) for binary classification.
Mdl = fitcgam (X, Y) returns a a GAM classification
model, Mdl, with X being the predictor data, and Y the
binary class labels of observations in X.
-
Xmust be a numeric matrix of predictor data where rows correspond to observations and columns correspond to features or variables. -
Yis numeric vector containing binary class labels, typically 0 or 1.
Mdl = fitcgam (…, name, value) returns a
GAM classification model with additional options specified by
Name-Value pair arguments listed below.
Model Parameters
| Name | Value | |
|---|---|---|
"PredictorNames" | A cell array of character vectors specifying the names of the predictors. The length of this array must match the number of columns in X. | |
"ResponseName" | A character vector specifying the name of the response variable. | |
"ClassNames" | Names of the classes in the class
labels, Y, used for fitting the Discriminant model. ClassNames
are of the same type as the class labels in Y. | |
"Cost" | A numeric matrix containing
misclassification cost for the corresponding instances in X where
is the number of unique categories in Y. If an instance is
correctly classified into its category the cost is calculated to be 1,
otherwise 0. cost matrix can be altered use Mdl.cost = somecost.
default value cost = ones(rows(X),numel(unique(Y))). | |
"Formula" | A model specification given as a string in
the form "Y ~ terms" where Y represents the reponse variable
and terms the predictor variables. The formula can be used to
specify a subset of variables for training model. For example:
"Y ~ x1 + x2 + x3 + x4 + x1:x2 + x2:x3" specifies four linear terms
for the first four columns of for predictor data, and x1:x2 and
x2:x3 specify the two interaction terms for 1st-2nd and 3rd-4th
columns respectively. Only these terms will be used for training the model,
but X must have at least as many columns as referenced in the formula.
If Predictor Variable names have been defined, then the terms in the formula
must reference to those. When "formula" is specified, all terms used
for training the model are referenced in the IntMatrix field of the
obj class object as a matrix containing the column indexes for each
term including both the predictors and the interactions used. | |
"Interactions" | A logical matrix, a positive integer
scalar, or the string "all" for defining the interactions between
predictor variables. When given a logical matrix, it must have the same
number of columns as X and each row corresponds to a different
interaction term combining the predictors indexed as true. Each
interaction term is appended as a column vector after the available predictor
column in X. When "all" is defined, then all possible
combinations of interactions are appended in X before training. At the
moment, parsing a positive integer has the same effect as the "all"
option. When "interactions" is specified, only the interaction terms
appended to X are referenced in the IntMatrix field of the
obj class object. | |
"Knots" | A scalar or a row vector with the same
columns as X. It defines the knots for fitting a polynomial when
training the GAM. As a scalar, it is expanded to a row vector. The default
value is 5, hence expanded to ones (1, columns (X)) * 5. You can
parse a row vector with different number of knots for each predictor
variable to be fitted with, although not recommended. | |
"Order" | A scalar or a row vector with the same
columns as X. It defines the order of the polynomial when training the
GAM. As a scalar, it is expanded to a row vector. The default values is 3,
hence expanded to ones (1, columns (X)) * 3. You can parse a row
vector with different number of polynomial order for each predictor variable
to be fitted with, although not recommended. | |
"DoF" | A scalar or a row vector with the same columns
as X. It defines the degrees of freedom for fitting a polynomial when
training the GAM. As a scalar, it is expanded to a row vector. The default
value is 8, hence expanded to ones (1, columns (X)) * 8. You can
parse a row vector with different degrees of freedom for each predictor
variable to be fitted with, although not recommended. |
You can parse either a "Formula" or an "Interactions"
optional parameter. Parsing both parameters will result an error.
Accordingly, you can only pass up to two parameters among "Knots",
"Order", and "DoF" to define the required polynomial for
training the GAM model.
See also: ClassificationGAM
Source Code: fitcgam
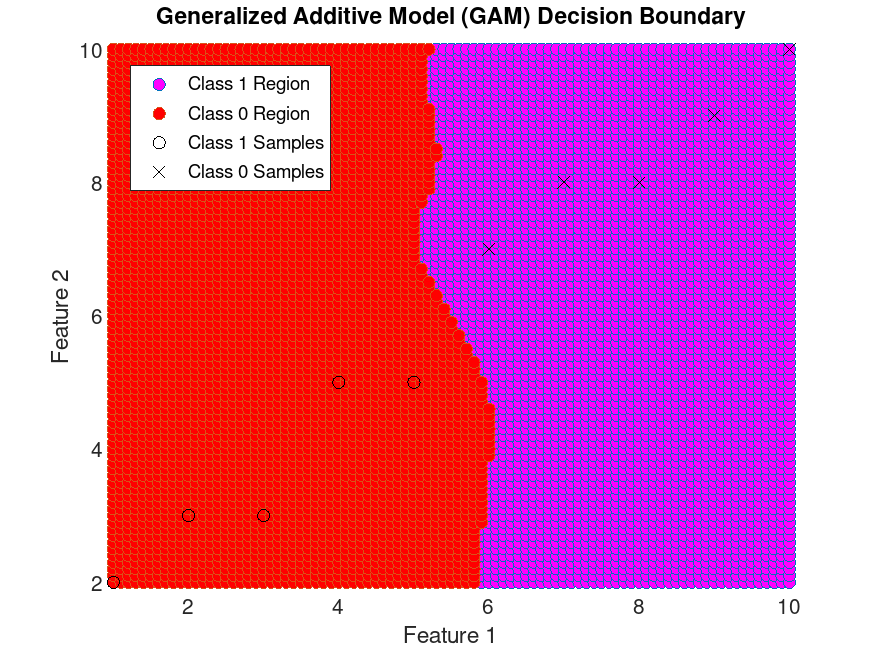
Example: 1
## Train a GAM classifier for binary classification
## using specific data and plot the decision boundaries.
## Define specific data
X = [1, 2; 2, 3; 3, 3; 4, 5; 5, 5; ...
6, 7; 7, 8; 8, 8; 9, 9; 10, 10];
Y = [0; 0; 0; 0; 0; ...
1; 1; 1; 1; 1];
## Train the GAM model
obj = fitcgam (X, Y, "Interactions", "all");
## Create a grid of values for prediction
x1 = [min(X(:,1)):0.1:max(X(:,1))];
x2 = [min(X(:,2)):0.1:max(X(:,2))];
[x1G, x2G] = meshgrid (x1, x2);
XGrid = [x1G(:), x2G(:)];
pred = predict (obj, XGrid);
## Plot decision boundaries and data points
predNumeric = str2double (pred);
gidx = predNumeric > 0.5;
figure
scatter(XGrid(gidx,1), XGrid(gidx,2), "markerfacecolor", "magenta");
hold on
scatter(XGrid(!gidx,1), XGrid(!gidx,2), "markerfacecolor", "red");
plot(X(Y == 0, 1), X(Y == 0, 2), "ko", X(Y == 1, 1), X(Y == 1, 2), "kx");
xlabel("Feature 1");
ylabel("Feature 2");
title("Generalized Additive Model (GAM) Decision Boundary");
legend({"Class 1 Region", "Class 0 Region", ...
"Class 1 Samples", "Class 0 Samples"}, ...
"location", "northwest")
axis tight
hold off
|