Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: bisafit
statistics: paramhat = bisafit (x)
statistics: [paramhat, paramci] = bisafit (x)
statistics: [paramhat, paramci] = bisafit (x, alpha)
statistics: […] = bisafit (x, alpha, censor)
statistics: […] = bisafit (x, alpha, censor, freq)
statistics: […] = bisafit (x, alpha, censor, freq, options)
Estimate mean and confidence intervals for the Birnbaum-Saunders distribution.
muhat = bisafit (x) returns the maximum likelihood
estimates of the parameters of the Birnbaum-Saunders distribution given the
data in x. paramhat(1) is the scale parameter,
beta, and paramhat(2) is the shape parameter,
gamma.
[paramhat, paramci] = bisafit (x) returns the 95%
confidence intervals for the parameter estimates.
[…] = bisafit (x, alpha) also returns the
100 * (1 - alpha) percent confidence intervals for the
parameter estimates. By default, the optional argument alpha is
0.05 corresponding to 95% confidence intervals. Pass in [] for
alpha to use the default values.
[…] = bisafit (x, alpha, censor) accepts a
boolean vector, censor, of the same size as x with 1s for
observations that are right-censored and 0s for observations that are
observed exactly. By default, or if left empty,
censor = zeros (size (x)).
[…] = bisafit (x, alpha, censor, freq)
accepts a frequency vector, freq, of the same size as x.
freq typically contains integer frequencies for the corresponding
elements in x, but it can contain any non-integer non-negative values.
By default, or if left empty, freq = ones (size (x)).
[…] = bisafit (…, options) specifies control
parameters for the iterative algorithm used to compute ML estimates with the
fminsearch function. options is a structure with the following
fields and their default values:
-
options.Display = "off" -
options.MaxFunEvals = 400 -
options.MaxIter = 200 -
options.TolX = 1e-6
Further information about the Birnbaum-Saunders distribution can be found at https://en.wikipedia.org/wiki/Birnbaum%E2%80%93Saunders_distribution
See also: bisacdf, bisainv, bisapdf, bisarnd, bisalike, bisastat
Source Code: bisafit
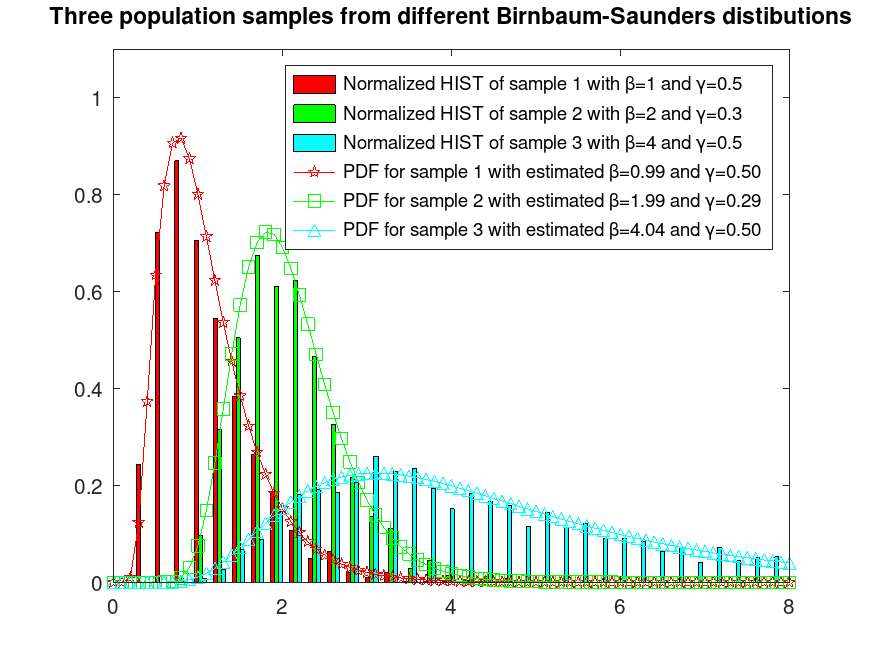
Example: 1
## Sample 3 populations from different Birnbaum-Saunders distributions
rand ("seed", 5); # for reproducibility
r1 = bisarnd (1, 0.5, 2000, 1);
rand ("seed", 2); # for reproducibility
r2 = bisarnd (2, 0.3, 2000, 1);
rand ("seed", 7); # for reproducibility
r3 = bisarnd (4, 0.5, 2000, 1);
r = [r1, r2, r3];
## Plot them normalized and fix their colors
hist (r, 80, 4.2);
h = findobj (gca, "Type", "patch");
set (h(1), "facecolor", "c");
set (h(2), "facecolor", "g");
set (h(3), "facecolor", "r");
ylim ([0, 1.1]);
xlim ([0, 8]);
hold on
## Estimate their α and β parameters
beta_gammaA = bisafit (r(:,1));
beta_gammaB = bisafit (r(:,2));
beta_gammaC = bisafit (r(:,3));
## Plot their estimated PDFs
x = [0:0.1:8];
y = bisapdf (x, beta_gammaA(1), beta_gammaA(2));
plot (x, y, "-pr");
y = bisapdf (x, beta_gammaB(1), beta_gammaB(2));
plot (x, y, "-sg");
y = bisapdf (x, beta_gammaC(1), beta_gammaC(2));
plot (x, y, "-^c");
hold off
legend ({"Normalized HIST of sample 1 with β=1 and γ=0.5", ...
"Normalized HIST of sample 2 with β=2 and γ=0.3", ...
"Normalized HIST of sample 3 with β=4 and γ=0.5", ...
sprintf("PDF for sample 1 with estimated β=%0.2f and γ=%0.2f", ...
beta_gammaA(1), beta_gammaA(2)), ...
sprintf("PDF for sample 2 with estimated β=%0.2f and γ=%0.2f", ...
beta_gammaB(1), beta_gammaB(2)), ...
sprintf("PDF for sample 3 with estimated β=%0.2f and γ=%0.2f", ...
beta_gammaC(1), beta_gammaC(2))})
title ("Three population samples from different Birnbaum-Saunders distributions")
hold off
|