Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Class Definition: tLocationScaleDistribution
statistics: tLocationScaleDistribution
Location-Scale Student’s T probability distribution object.
A tLocationScaleDistribution object consists of parameters, a model
description, and sample data for a location-scale Student’s T probability
distribution.
The location-scale Student’s T distribution is a continuous probability distribution that generalizes the standard Student’s T distribution by including location and scale parameters. It is defined by location parameter mu, scale parameter sigma, and degrees of freedom nu.
There are several ways to create a tLocationScaleDistribution object.
- Fit a distribution to data using the
fitdistfunction. - Create a distribution with fixed parameter values using the
makedistfunction. - Use the constructor
tLocationScaleDistribution (mu, sigma, nu)to create a location-scale Student’s T distribution with fixed parameter values mu, sigma, and nu. - Use the static method
tLocationScaleDistribution.fit (x, censor, freq, options)to fit a distribution to the data in x using the same input arguments as thetlsfitfunction.
It is highly recommended to use fitdist and makedist
functions to create probability distribution objects, instead of the class
constructor or the aforementioned static method.
Further information about the location-scale Student’s T distribution can be found at https://en.wikipedia.org/wiki/Student%27s_t-distribution#Location-scale_t_distribution
See also: fitdist, makedist, tlscdf, tlsinv, tlspdf, tlsrnd, tlsfit, tlslike, tlsstat
Source Code: tLocationScaleDistribution
Properties
A scalar value characterizing the location of the
location-scale Student’s T distribution. You can access the mu
property using dot name assignment.
Example: 1
## Create a t Location-Scale distribution by fitting to data
data = tlsrnd (0, 1, 5, 10000, 1); % Generate data with mu=0, sigma=1, nu=5
pd = fitdist (data, "tLocationScale");
## Query parameter 'mu' (location parameter)
pd.mu
## Set parameter 'mu'
pd.mu = 1
## Use this to initialize or modify the location parameter of a t Location-Scale
## distribution. The location parameter (mu) is a real scalar that shifts the
## distribution, useful for modeling data centered around a specific value.
ans = -0.012491
pd =
tLocationScaleDistribution
t Location-Scale distribution
mu = 1
sigma = 0.997056
nu = 4.98394
|
Example: 2
## Create a t Location-Scale distribution object by calling its constructor
pd = tLocationScaleDistribution (2, 1, 5);
## Query parameter 'mu'
pd.mu
## This demonstrates direct construction with a specific location parameter,
## ideal for modeling data with a known center, such as test scores or residuals.
ans = 2
|
A positive scalar value characterizing the scale of the
location-scale Student’s T distribution. You can access the sigma
property using dot name assignment.
Example: 1
## Create a t Location-Scale distribution with fitted parameters
data = tlsrnd (0, 1, 5, 10000, 1); % Generate data with mu=0, sigma=1, nu=5
pd = fitdist (data, "tLocationScale");
## Query parameter 'sigma' (scale parameter)
pd.sigma
## Set parameter 'sigma'
pd.sigma = 2
## Use this to initialize or modify the scale parameter, which controls the
## spread of the t Location-Scale distribution. Sigma must be a positive real
## scalar, useful for modeling variability in data like financial returns.
ans = 0.9880
pd =
tLocationScaleDistribution
t Location-Scale distribution
mu = -0.0154656
sigma = 2
nu = 5.16791
|
Example: 2
## Create a t Location-Scale distribution object by calling its constructor
pd = tLocationScaleDistribution (0, 1.5, 5);
## Query parameter 'sigma'
pd.sigma
## This shows how to set the scale parameter directly via the constructor,
## useful for modeling data with specific variability, such as process errors.
ans = 1.5000
|
A positive scalar value characterizing the degrees of freedom of the
location-scale Student’s T distribution. You can access the nu
property using dot name assignment.
Example: 1
## Create a t Location-Scale distribution with fitted parameters
data = tlsrnd (0, 1, 5, 10000, 1); % Generate data with mu=0, sigma=1, nu=5
pd = fitdist (data, "tLocationScale");
## Query parameter 'nu' (degrees of freedom)
pd.nu
## Set parameter 'nu'
pd.nu = 10
## Use this to initialize or modify the degrees of freedom, which controls the
## tail heaviness of the t Location-Scale distribution. Nu must be a positive
## real scalar, useful for modeling heavy-tailed data like stock returns.
ans = 4.8489
pd =
tLocationScaleDistribution
t Location-Scale distribution
mu = 0.0126855
sigma = 1.00002
nu = 10
|
Example: 2
## Create a t Location-Scale distribution object by calling its constructor
pd = tLocationScaleDistribution (0, 1, 3);
## Query parameter 'nu'
pd.nu
## This demonstrates setting the degrees of freedom directly via the constructor,
## ideal for modeling data with specific tail behavior, such as outlier-prone datasets.
ans = 3
|
A character vector specifying the name of the probability distribution object. This property is read-only.
A scalar integer value specifying the number of parameters characterizing the probability distribution. This property is read-only.
A cell array of character vectors with each element containing the name of a distribution parameter. This property is read-only.
A cell array of character vectors with each element containing a short description of a distribution parameter. This property is read-only.
A numeric vector containing the values of the distribution
parameters. This property is read-only. You can change the distribution
parameters by assigning new values to the mu, sigma, and
nu properties.
A numeric matrix containing the variance-covariance of the parameter estimates. Diagonal elements contain the variance of each estimated parameter, and non-diagonal elements contain the covariance between the parameter estimates. The covariance matrix is only meaningful when the distribution was fitted to data. If the distribution object was created with fixed parameters, or a parameter of a fitted distribution is modified, then all elements of the variance-covariance are zero. This property is read-only.
A logical vector specifying which parameters are fixed and
which are estimated. true values correspond to fixed parameters,
false values correspond to parameter estimates. This property is
read-only.
A numeric vector specifying the truncation interval for the
probability distribution. First element contains the lower boundary,
second element contains the upper boundary. This property is read-only.
You can only truncate a probability distribution with the
truncate method.
A logical scalar value specifying whether a probability distribution is truncated or not. This property is read-only.
A scalar structure containing the following fields:
-
data: a numeric vector containing the data used for distribution fitting. -
cens: a numeric vector of logical values indicating censoring information corresponding to the elements of the data used for distribution fitting. If no censoring vector was used for distribution fitting, then this field defaults to an empty array. -
freq: a numeric vector of non-negative integer values containing the frequency information corresponding to the elements of the data used for distribution fitting. If no frequency vector was used for distribution fitting, then this field defaults to an empty array.
Methods
tLocationScaleDistribution: p = cdf (pd, x)
tLocationScaleDistribution: p = cdf (pd, x,
"upper")
p = cdf (pd, x) computes the CDF of the
probability distribution object, pd, evaluated at the values in
x.
p = cdf (…, returns the complement of
the CDF of the probability distribution object, pd, evaluated at
the values in x.
"upper")
Example: 1
## Plot various CDFs from the t Location-Scale distribution
x = -5:0.01:5;
data1 = tlsrnd (0, 0.5, 5, 10000, 1);
data2 = tlsrnd (0, 1, 5, 10000, 1);
data3 = tlsrnd (0, 2, 5, 10000, 1);
pd1 = fitdist (data1, "tLocationScale");
pd2 = fitdist (data2, "tLocationScale");
pd3 = fitdist (data3, "tLocationScale");
p1 = cdf (pd1, x);
p2 = cdf (pd2, x);
p3 = cdf (pd3, x);
plot (x, p1, "-b", x, p2, "-g", x, p3, "-r")
grid on
legend ({"mu = 0, sigma = 0.5, nu = 5", "mu = 0, sigma = 1, nu = 5", ...
"mu = 0, sigma = 2, nu = 5"}, "location", "southeast")
title ("t Location-Scale CDF")
xlabel ("Values in x")
ylabel ("Cumulative probability")
## Use this to compute and visualize the cumulative distribution function
## for different t Location-Scale distributions, showing how probability
## accumulates, useful in risk analysis or hypothesis testing.
|
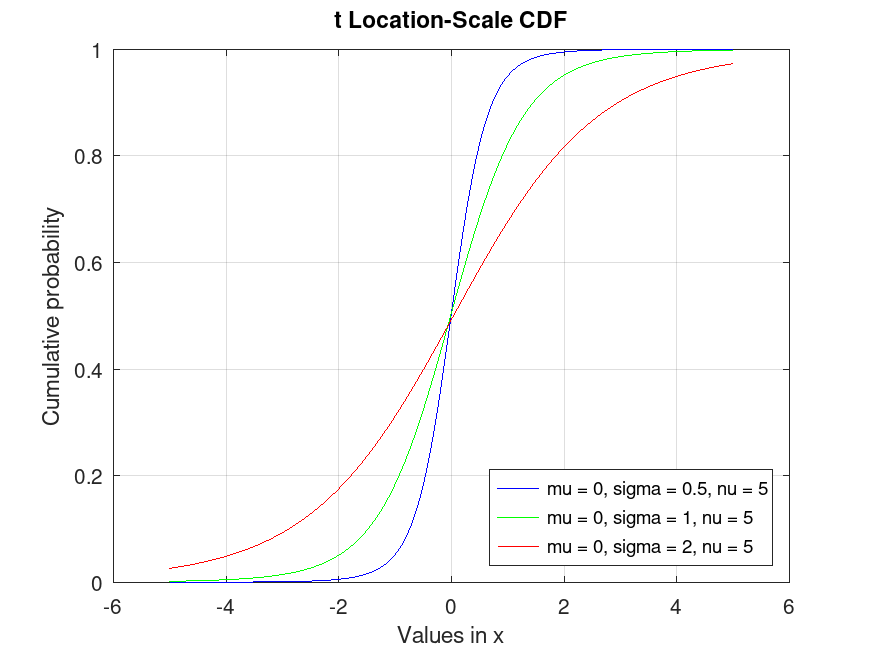
tLocationScaleDistribution: x = icdf (pd, p)
x = icdf (pd, p) computes the quantile (the
inverse of the CDF) of the probability distribution object, pd,
evaluated at the values in p.
Example: 1
## Plot various iCDFs from the t Location-Scale distribution
p = 0.001:0.001:0.999;
data1 = tlsrnd (0, 0.5, 5, 10000, 1);
data2 = tlsrnd (0, 1, 5, 10000, 1);
data3 = tlsrnd (0, 2, 5, 10000, 1);
pd1 = fitdist (data1, "tLocationScale");
pd2 = fitdist (data2, "tLocationScale");
pd3 = fitdist (data3, "tLocationScale");
x1 = icdf (pd1, p);
x2 = icdf (pd2, p);
x3 = icdf (pd3, p);
plot (p, x1, "-b", p, x2, "-g", p, x3, "-r")
grid on
legend ({"mu = 0, sigma = 0.5, nu = 5", "mu = 0, sigma = 1, nu = 5", ...
"mu = 0, sigma = 2, nu = 5"}, "location", "northwest")
title ("t Location-Scale iCDF")
xlabel ("Probability")
ylabel ("Values in x")
## This demonstrates the inverse CDF (quantiles) for t Location-Scale
## distributions, useful for finding critical values or thresholds in statistical testing.
|
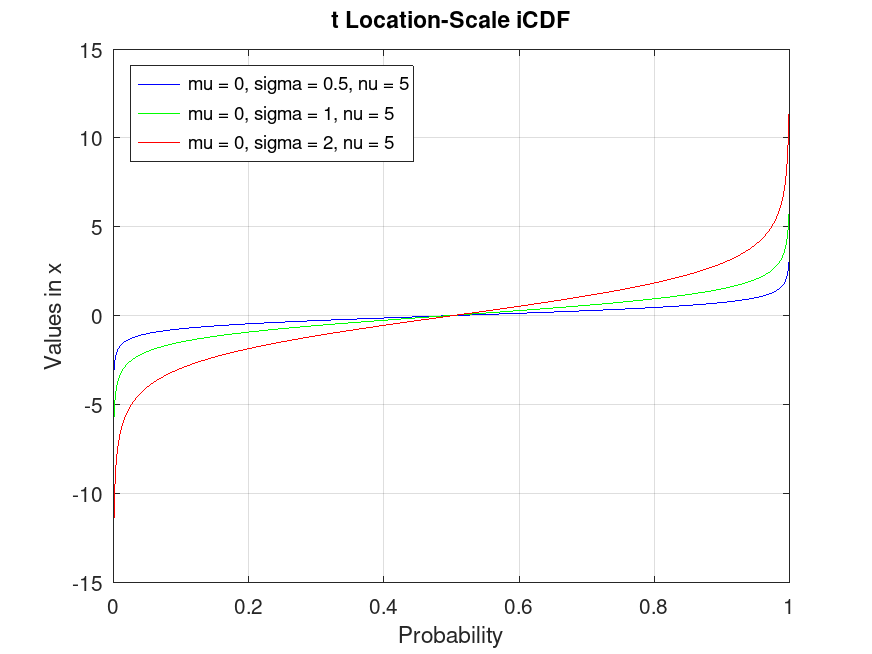
tLocationScaleDistribution: r = iqr (pd)
r = iqr (pd) computes the interquartile range of the
probability distribution object, pd.
Example: 1
## Compute the interquartile range for a t Location-Scale distribution
data = tlsrnd (0, 1, 5, 10000, 1);
pd = fitdist (data, "tLocationScale");
iqr_value = iqr (pd)
## Use this to calculate the interquartile range, which measures the spread
## of the middle 50% of the distribution, helpful for assessing central variability.
iqr_value = 1.4420
|
tLocationScaleDistribution: m = mean (pd)
m = mean (pd) computes the mean of the probability
distribution object, pd.
Example: 1
## Compute the mean for different t Location-Scale distributions
data1 = tlsrnd (0, 0.5, 5, 10000, 1);
data2 = tlsrnd (0, 1, 5, 10000, 1);
pd1 = fitdist (data1, "tLocationScale");
pd2 = fitdist (data2, "tLocationScale");
mean1 = mean (pd1)
mean2 = mean (pd2)
## This shows how to compute the expected value for t Location-Scale
## distributions, representing the average value, useful in financial modeling or quality control.
mean1 = 1.1194e-03
mean2 = -0.012212
|
tLocationScaleDistribution: m = median (pd)
m = median (pd) computes the median of the probability
distribution object, pd.
Example: 1
## Compute the median for different t Location-Scale distributions
data1 = tlsrnd (0, 0.5, 5, 10000, 1);
data2 = tlsrnd (0, 1, 5, 10000, 1);
pd1 = fitdist (data1, "tLocationScale");
pd2 = fitdist (data2, "tLocationScale");
median1 = median (pd1)
median2 = median (pd2)
## Use this to find the median value, which splits the distribution into
## two equal probability halves, robust to heavy-tailed data.
median1 = 1.0982e-03
median2 = 0.017472
|
tLocationScaleDistribution: nlogL = negloglik (pd)
nlogL = negloglik (pd) computes the negative
loglikelihood of the probability distribution object, pd.
Example: 1
## Compute the negative loglikelihood for a fitted t Location-Scale distribution
rand ("seed", 21);
data = tlsrnd (0, 1, 5, 100, 1);
pd_fitted = fitdist (data, "tLocationScale");
params = [pd_fitted.mu, pd_fitted.sigma, pd_fitted.nu];
nlogL_tlslike = tlslike (params, data)
## This is useful for assessing the fit of a t Location-Scale distribution to
## data, with lower values indicating a better fit, often used in model comparison.
nlogL_tlslike = 176.78
|
tLocationScaleDistribution: ci = paramci (pd)
tLocationScaleDistribution: ci = paramci (pd, Name, Value)
ci = paramci (pd) computes the lower and upper
boundaries of the 95% confidence interval for each parameter of the
probability distribution object, pd.
ci = paramci (pd, Name, Value) computes
the confidence intervals with additional options specified by
Name-Value pair arguments listed below.
| Name | Value | |
|---|---|---|
"Alpha" | A scalar value in the range specifying the significance level for the confidence interval. The default value 0.05 corresponds to a 95% confidence interval. | |
"Parameter" | A character vector or a cell array of
character vectors specifying the parameter names for which to compute
confidence intervals. By default, paramci computes confidence
intervals for all distribution parameters. |
paramci is meaningful only when pd is fitted to data,
otherwise an empty array, [], is returned.
Example: 1
## Compute confidence intervals for parameters of a fitted t Location-Scale
## distribution
rand ("seed", 21);
data = tlsrnd (0, 1, 5, 1000, 1);
pd_fitted = fitdist (data, "tLocationScale");
ci = paramci (pd_fitted, "Alpha", 0.05)
## Use this to obtain confidence intervals for the estimated parameters (mu,
## sigma, nu), providing a range of plausible values given the data.
ci =
-0.060216 0.981854 3.618273
0.091896 1.137822 6.434089
|
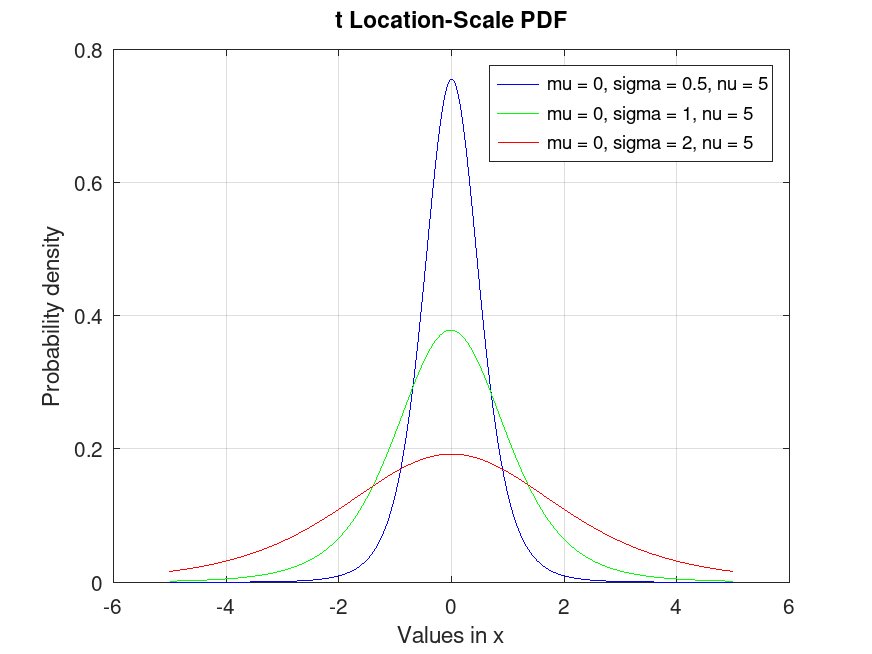
tLocationScaleDistribution: y = pdf (pd, x)
y = pdf (pd, x) computes the PDF of the
probability distribution object, pd, evaluated at the values in
x.
Example: 1
## Plot various PDFs from the t Location-Scale distribution
x = -5:0.01:5;
data1 = tlsrnd (0, 0.5, 5, 10000, 1);
data2 = tlsrnd (0, 1, 5, 10000, 1);
data3 = tlsrnd (0, 2, 5, 10000, 1);
pd1 = fitdist (data1, "tLocationScale");
pd2 = fitdist (data2, "tLocationScale");
pd3 = fitdist (data3, "tLocationScale");
y1 = pdf (pd1, x);
y2 = pdf (pd2, x);
y3 = pdf (pd3, x);
plot (x, y1, "-b", x, y2, "-g", x, y3, "-r")
grid on
legend ({"mu = 0, sigma = 0.5, nu = 5", "mu = 0, sigma = 1, nu = 5", ...
"mu = 0, sigma = 2, nu = 5"}, "location", "northeast")
title ("t Location-Scale PDF")
xlabel ("Values in x")
ylabel ("Probability density")
## This visualizes the probability density function for t Location-Scale
## distributions, showing the likelihood across values, useful for data analysis.
|
tLocationScaleDistribution: plot (pd)
tLocationScaleDistribution: plot (pd, Name, Value)
tLocationScaleDistribution: h = plot (…)
plot (pd) plots a probability density function (PDF) of the
probability distribution object pd. If pd contains data,
which have been fitted by fitdist, the PDF is superimposed over a
histogram of the data.
plot (pd, Name, Value) specifies additional
options with the Name-Value pair arguments listed below.
| Name | Value | |
|---|---|---|
"PlotType" | A character vector specifying the plot
type. "pdf" plots the probability density function (PDF). When
pd is fit to data, the PDF is superimposed on a histogram of the
data. "cdf" plots the cumulative density function (CDF). When
pd is fit to data, the CDF is superimposed over an empirical CDF.
"probability" plots a probability plot using a CDF of the data
and a CDF of the fitted probability distribution. This option is
available only when pd is fitted to data. | |
"Discrete" | A logical scalar to specify whether to
plot the PDF or CDF of a discrete distribution object as a line plot or a
stem plot, by specifying false or true, respectively. By
default, it is true for discrete distributions and false
for continuous distributions. When pd is a continuous distribution
object, option is ignored. | |
"Parent" | An axes graphics object for plot. If
not specified, the plot function plots into the current axes or
creates a new axes object if one does not exist. |
h = plot (…) returns a graphics handle to the plotted
objects.
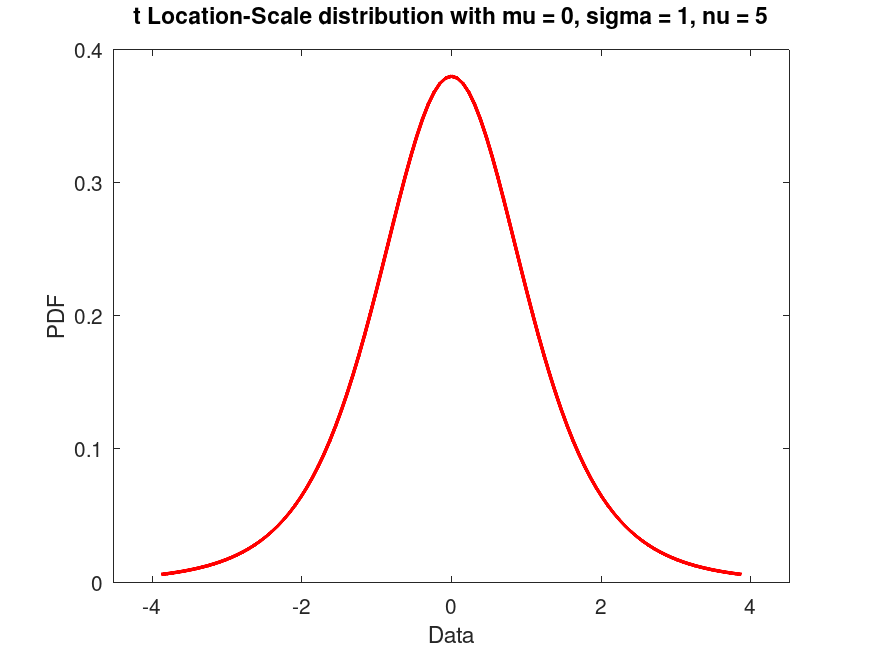
Example: 1
## Create a t Location-Scale distribution with fixed parameters and plot its PDF
pd = tLocationScaleDistribution (0, 1, 5);
plot (pd)
title ("t Location-Scale distribution with mu = 0, sigma = 1, nu = 5")
## Use this to visualize the PDF of a t Location-Scale distribution with
## fixed parameters, helpful for theoretical exploration.
|
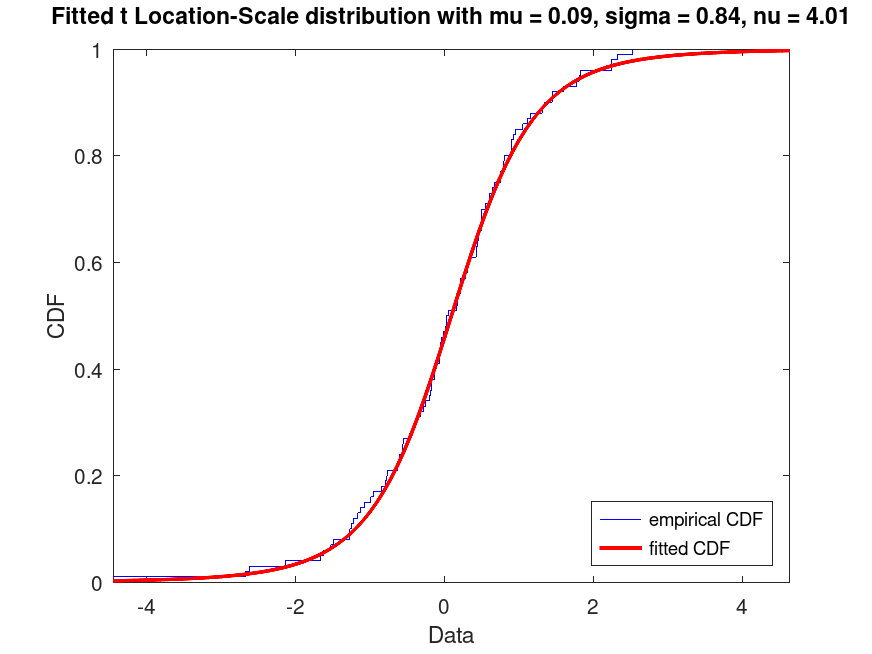
Example: 2
## Generate a data set and plot the CDF of a fitted t Location-Scale distribution
rand ("seed", 21);
data = tlsrnd (0, 1, 5, 100, 1);
pd_fitted = fitdist (data, "tLocationScale");
plot (pd_fitted, "PlotType", "cdf")
txt = "Fitted t Location-Scale distribution with mu = %0.2f, sigma = %0.2f, nu = %0.2f";
title (sprintf (txt, pd_fitted.mu, pd_fitted.sigma, pd_fitted.nu))
legend ({"empirical CDF", "fitted CDF"}, "location", "southeast")
## Use this to visualize the fitted CDF compared to the empirical CDF of the
## data, useful for assessing model fit.
|
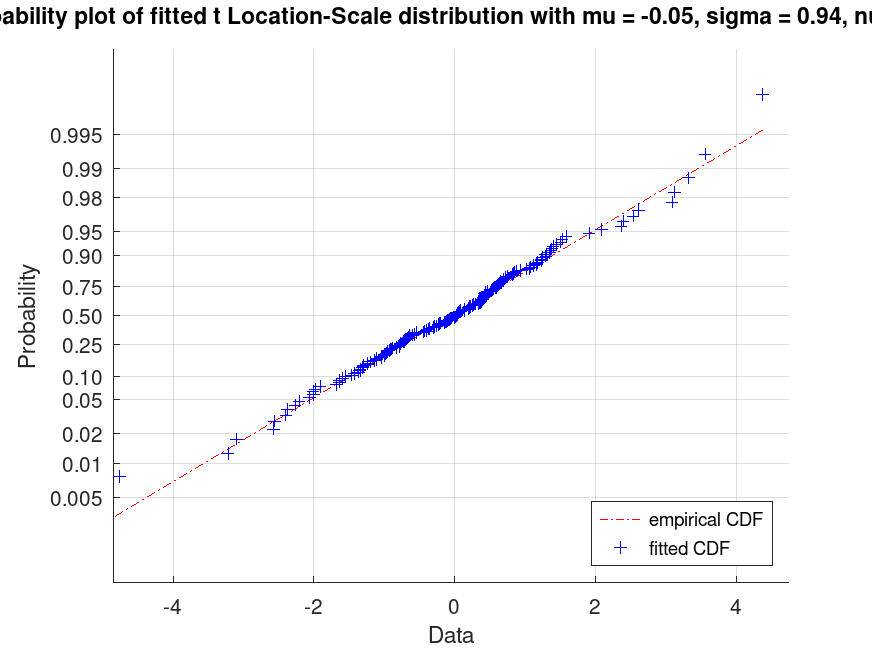
Example: 3
## Generate a data set and display a probability plot for a fitted t Location-Scale distribution
rand ("seed", 21);
data = tlsrnd (0, 1, 5, 200, 1);
pd_fitted = fitdist (data, "tLocationScale");
plot (pd_fitted, "PlotType", "probability")
txt = strcat ("Probability plot of fitted t Location-Scale", ...
" distribution with mu = %0.2f, sigma = %0.2f, nu = %0.2f");
title (sprintf (txt, pd_fitted.mu, pd_fitted.sigma, pd_fitted.nu))
legend ({"empirical CDF", "fitted CDF"}, "location", "southeast")
## This creates a probability plot to compare the fitted distribution to the
## data, useful for checking if the t Location-Scale model is appropriate.
|
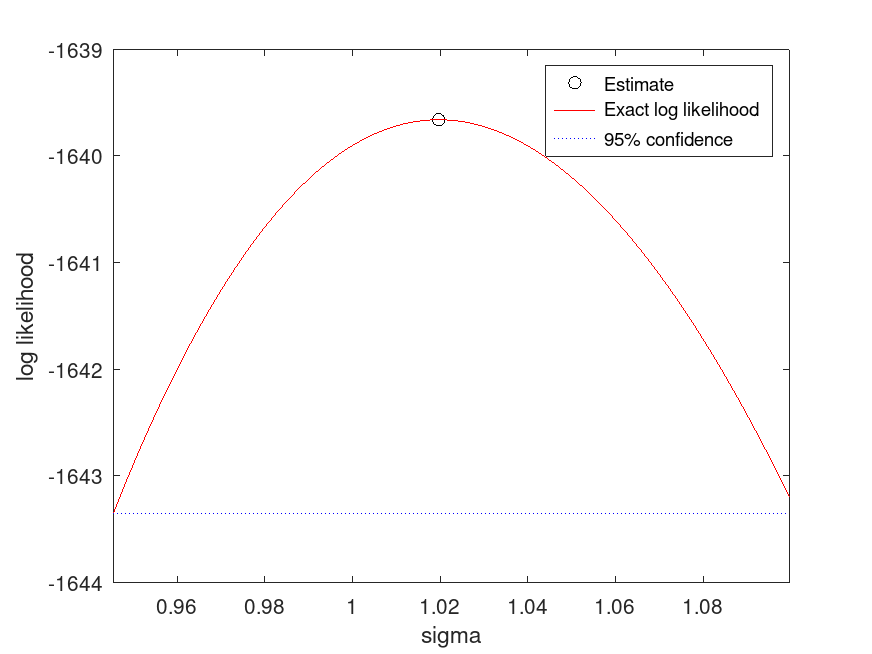
tLocationScaleDistribution: [nlogL, param] = proflik (pd, pnum)
tLocationScaleDistribution: [nlogL, param] = proflik (pd, pnum,
"Display", display)tLocationScaleDistribution: [nlogL, param] = proflik (pd, pnum, setparam)
tLocationScaleDistribution: [nlogL, param] = proflik (pd, pnum, setparam,
"Display", display)
[nlogL, param] = proflik (pd, pnum)
returns a vector nlogL of negative loglikelihood values and a
vector param of corresponding parameter values for the parameter in
the position indicated by pnum. By default, proflik uses
the lower and upper bounds of the 95% confidence interval and computes
100 equispaced values for the selected parameter. pd must be
fitted to data.
[nlogL, param] = proflik (pd, pnum,
also plots the profile likelihood
against the default range of the selected parameter.
"Display", "on")
[nlogL, param] = proflik (pd, pnum,
setparam) defines a user-defined range of the selected parameter.
[nlogL, param] = proflik (pd, pnum,
setparam, also plots the profile
likelihood against the user-defined range of the selected parameter.
"Display", "on")
For the location-scale Student’s T distribution, pnum = 1
selects the parameter mu, pnum = 2 selects the
parameter sigma, and pnum = 3 selects the
parameter nu.
When opted to display the profile likelihood plot, proflik also
plots the baseline loglikelihood computed at the lower bound of the 95%
confidence interval and estimated maximum likelihood. The latter might
not be observable if it is outside of the used-defined range of parameter
values.
Example: 1
## Compute and plot the profile likelihood for the scale parameter
rand ("seed", 21);
data = tlsrnd (0, 1, 5, 1000, 1);
pd_fitted = fitdist (data, "tLocationScale");
[nlogL, param] = proflik (pd_fitted, 2, "Display", "on");
## Use this to analyze the profile likelihood of the scale parameter (sigma),
## helping to understand the uncertainty in parameter estimates.
|
tLocationScaleDistribution: r = random (pd)
tLocationScaleDistribution: r = random (pd, rows)
tLocationScaleDistribution: r = random (pd, rows, cols, …)
tLocationScaleDistribution: r = random (pd, [sz])
r = random (pd) returns a random number from the
distribution object pd.
When called with a single size argument, tlsrnd returns a square
matrix with the dimension specified. When called with more than one
scalar argument, the first two arguments are taken as the number of rows
and columns and any further arguments specify additional matrix
dimensions. The size may also be specified with a row vector of
dimensions, sz.
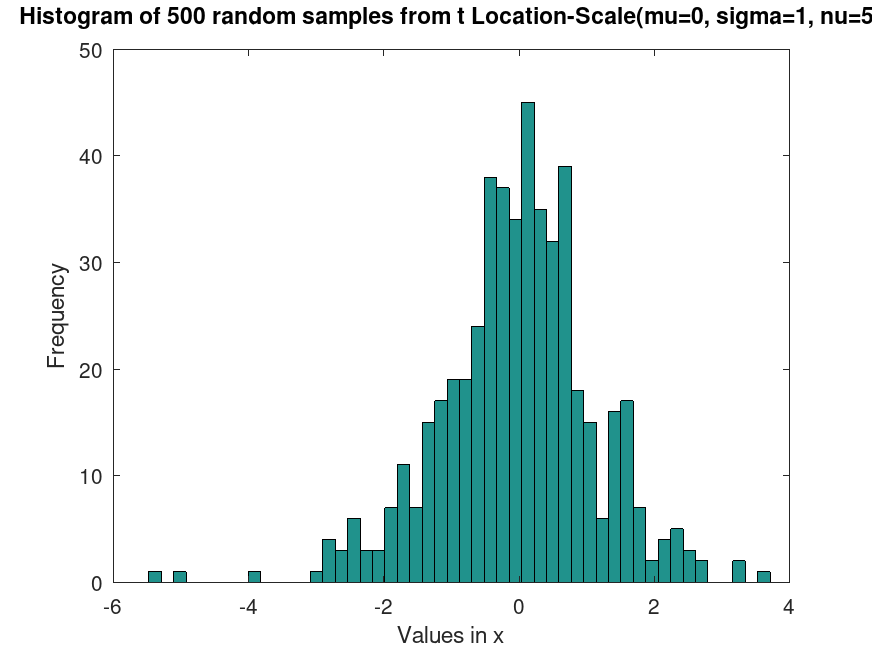
Example: 1
## Generate random samples from a t Location-Scale distribution
rand ("seed", 21);
samples = tlsrnd (0, 1, 5, 500, 1);
hist (samples, 50)
title ("Histogram of 500 random samples from t Location-Scale(mu=0, sigma=1, nu=5)")
xlabel ("Values in x")
ylabel ("Frequency")
## This generates random samples from a t Location-Scale distribution, useful
## for simulating data with heavy tails, such as financial or experimental data.
|
tLocationScaleDistribution: s = std (pd)
s = std (pd) computes the standard deviation of the
probability distribution object, pd.
Example: 1
## Compute the standard deviation for a t Location-Scale distribution
data = tlsrnd (0, 1, 5, 10000, 1);
pd = fitdist (data, "tLocationScale");
std_value = std (pd)
## Use this to calculate the standard deviation, which measures the variability
## in the distribution, useful for understanding data dispersion.
std_value = 1.2936
|
tLocationScaleDistribution: t = truncate (pd, lower, upper)
t = truncate (pd, lower, upper) returns a
probability distribution t, which is the probability distribution
pd truncated to the specified interval with lower limit,
lower, and upper limit, upper. If pd is fitted to data
with fitdist, the returned probability distribution t is not
fitted, does not contain any data or estimated values, and it is as it
has been created with the makedist function, but it includes the
truncation interval.
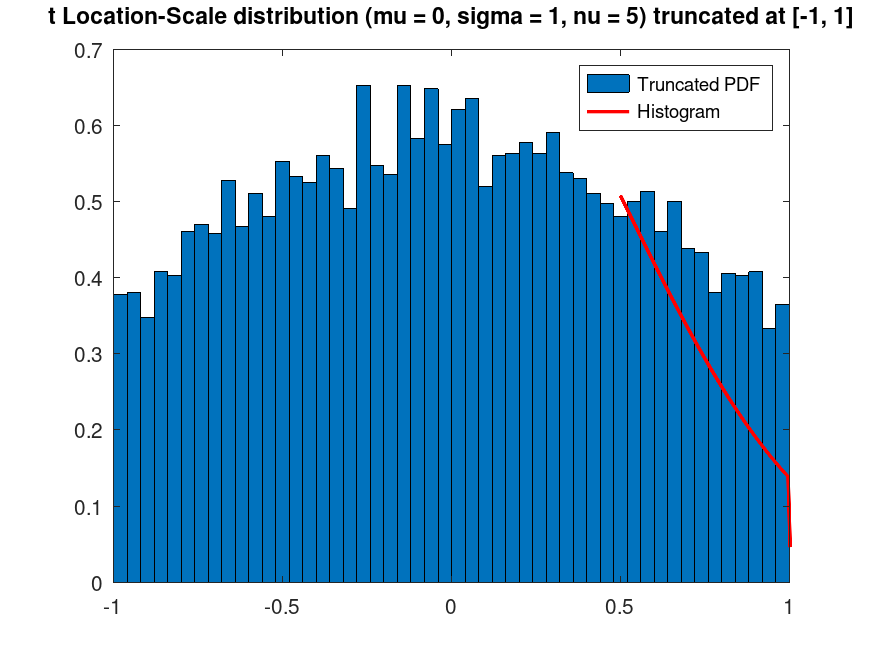
Example: 1
## Plot the PDF of a truncated t Location-Scale distribution
rand ("seed", 21);
data_all = tlsrnd (0, 1, 5, 20000, 1);
data = data_all(data_all >= -1 & data_all <= 1);
data = data(1:10000);
pd = fitdist (data, "tLocationScale");
t = truncate (pd, -1, 1);
[counts, centers] = hist (data, 50);
bin_width = centers(2) - centers(1);
bar (centers, counts / (sum (counts) * bin_width), 1);
hold on;
## Plot histogram and truncated PDF
x = linspace (0.5, 5, 500);
y = pdf (t, x);
plot (x, y, "r", "linewidth", 2);
title ("t Location-Scale distribution (mu = 0, sigma = 1, nu = 5) truncated at [-1, 1]")
legend ("Truncated PDF", "Histogram")
## This demonstrates truncating a t Location-Scale distribution to a specific
## range and visualizing the resulting distribution with random samples.
|
tLocationScaleDistribution: v = var (pd)
v = var (pd) computes the variance of the
probability distribution object, pd.
Example: 1
## Compute the variance for a t Location-Scale distribution
data = tlsrnd (0, 1, 5, 10000, 1);
pd = fitdist (data, "tLocationScale");
var_value = var (pd)
## Use this to calculate the variance, which quantifies the spread of the
## distribution, useful for statistical analysis of variability.
var_value = 1.6678
|
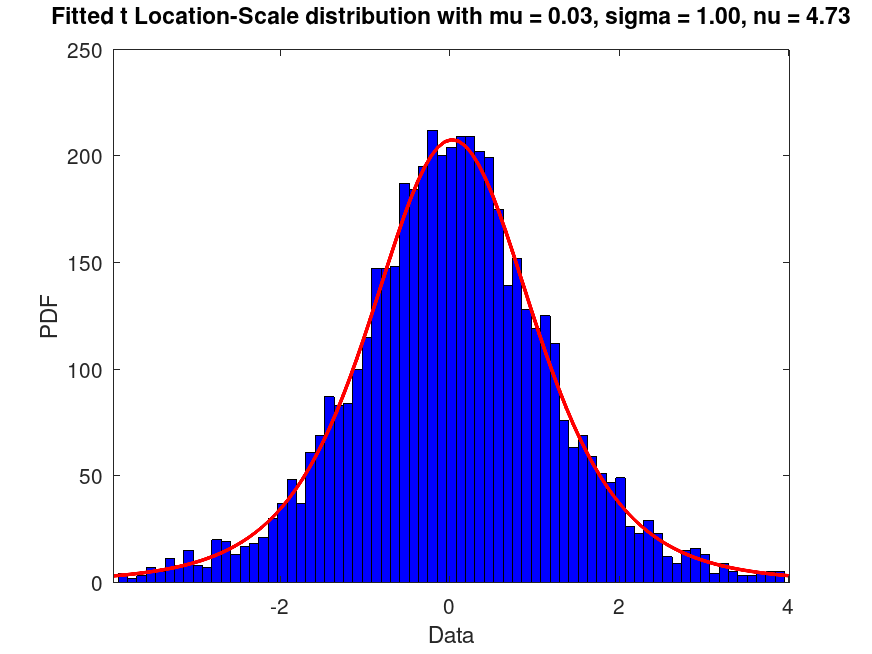
Examples
pd_fixed = makedist ("tLocationScale", "mu", 0, "sigma", 1, "nu", 5);
rand ("seed", 2);
data = random (pd_fixed, 5000, 1);
pd_fitted = fitdist (data, "tLocationScale");
plot (pd_fitted);
msg = "Fitted t Location-Scale distribution with mu = %0.2f, sigma = %0.2f, nu = %0.2f";
title (sprintf (msg, pd_fitted.mu, pd_fitted.sigma, pd_fitted.nu)); |