Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: fitcdiscr
statistics: Mdl = fitcdiscr (X, Y)
statistics: Mdl = fitcdiscr (…, name, value)
Fit a Linear Discriminant Analysis classification model.
Mdl = fitcdiscr (X, Y) returns a Linear Discriminant
Analysis (LDA) classification model, Mdl, with X being the
predictor data, and Y the class labels of observations in X.
-
Xmust be a numeric matrix of predictor data where rows correspond to observations and columns correspond to features or variables. -
Yis matrix or cell matrix containing the class labels of corresponding predictor data in X. Y can be numerical, logical, char array or cell array of character vectors. Y must have same number of rows as X.
Mdl = fitcdiscr (…, name, value) returns a
Linear Discriminant Analysis model with additional options specified by
Name-Value pair arguments listed below.
Model Parameters
| Name | Value | |
|---|---|---|
"PredictorNames" | A cell array of character vectors specifying the names of the predictors. The length of this array must match the number of columns in X. | |
"ResponseName" | A character vector specifying the name of the response variable. | |
"ClassNames" | Names of the classes in the class
labels, Y, used for fitting the Discriminant model. ClassNames
are of the same type as the class labels in Y. | |
"Prior" | A numeric vector specifying the prior
probabilities for each class. The order of the elements in Prior
corresponds to the order of the classes in ClassNames.
Alternatively, you can specify "empirical" to use the empirical
class probabilities or "uniform" to assume equal class probabilities. | |
"Cost" | A numeric matrix containing
misclassification cost for the corresponding instances in X where
is the number of unique categories in Y. If an instance is
correctly classified into its category the cost is calculated to be 1,
otherwise 0. cost matrix can be altered use Mdl.cost = somecost.
default value cost = ones(rows(X),numel(unique(Y))). | |
"DiscrimType" | A character vector or string scalar
specifying the type of discriminant analysis to perform. The only supported
value is "linear". | |
"FillCoeffs" | A character vector or string scalar
with values "on" or "off" specifying whether to fill the
coefficients after fitting. If set to "on", the coefficients are
computed during model fitting, which can be useful for prediction. | |
"Gamma" | A numeric scalar specifying the regularization parameter for the covariance matrix. It adjusts the linear discriminant analysis to make the model more stable in the presence of multicollinearity or small sample sizes. A value of 0 corresponds to no regularization, while a value of 1 corresponds to a completely regularized model. |
See also: ClassificationDiscriminant
Source Code: fitcdiscr
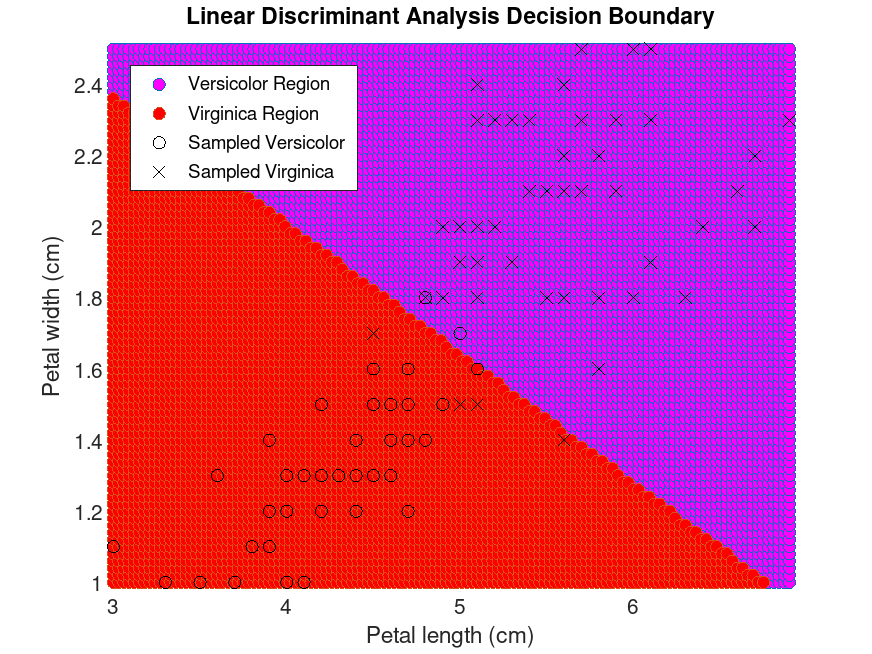
Example: 1
## Train a linear discriminant classifier for Gamma = 0.5
## and plot the decision boundaries.
load fisheriris
idx = ! strcmp (species, "setosa");
X = meas(idx,3:4);
Y = cast (strcmpi (species(idx), "virginica"), "double");
obj = fitcdiscr (X, Y, "Gamma", 0.5)
x1 = [min(X(:,1)):0.03:max(X(:,1))];
x2 = [min(X(:,2)):0.02:max(X(:,2))];
[x1G, x2G] = meshgrid (x1, x2);
XGrid = [x1G(:), x2G(:)];
pred = predict (obj, XGrid);
gidx = logical (pred);
figure
scatter (XGrid(gidx,1), XGrid(gidx,2), "markerfacecolor", "magenta");
hold on
scatter (XGrid(!gidx,1), XGrid(!gidx,2), "markerfacecolor", "red");
plot (X(Y == 0, 1), X(Y == 0, 2), "ko", X(Y == 1, 1), X(Y == 1, 2), "kx");
xlabel ("Petal length (cm)");
ylabel ("Petal width (cm)");
title ("Linear Discriminant Analysis Decision Boundary");
legend ({"Versicolor Region", "Virginica Region", ...
"Sampled Versicolor", "Sampled Virginica"}, ...
"location", "northwest")
axis tight
hold off
obj =
ClassificationDiscriminant
ResponseName: 'Y'
ClassNames: [0 1]
ScoreTransform: 'none'
NumObservations: 100
NumPredictors: 2
DiscrimType: 'linear'
Mu: [2x2 double]
Coeffs: [2x2 struct]
|