Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Class Definition: RegressionGAM
statistics: RegressionGAM
Create a RegressionGAM class object containing a Generalized Additive
Model (GAM) for regression.
A RegressionGAM class object can store the predictors and response
data along with various parameters for the GAM model. It is recommended to
use the fitrgam function to create a RegressionGAM object.
obj = RegressionGAM (X, Y) returns an object of
class RegressionGAM, with matrix X containing the predictor data and
vector Y containing the continuous response data.
- X must be a numeric matrix of input data where rows correspond to observations and columns correspond to features or variables. X will be used to train the GAM model.
- Y must be numeric vector containing the response data corresponding to the predictor data in X. Y must have same number of rows as X.
obj = RegressionGAM (…, name, value) returns
an object of class RegressionGAM with additional properties specified by
Name-Value pair arguments listed below.
| Name | Value | |
|---|---|---|
"predictors" | Predictor Variable names, specified as
a row vector cell of strings with the same length as the columns in X.
If omitted, the program will generate default variable names
(x1, x2, ..., xn) for each column in X. | |
"responsename" | Response Variable Name, specified as
a string. If omitted, the default value is "Y". | |
"formula" | a model specification given as a string in
the form "Y ~ terms" where Y represents the response variable
and terms the predictor variables. The formula can be used to
specify a subset of variables for training model. For example:
"Y ~ x1 + x2 + x3 + x4 + x1:x2 + x2:x3" specifies four linear terms
for the first four columns of for predictor data, and x1:x2 and
x2:x3 specify the two interaction terms for 1st-2nd and 3rd-4th
columns respectively. Only these terms will be used for training the model,
but X must have at least as many columns as referenced in the formula.
If Predictor Variable names have been defined, then the terms in the formula
must reference to those. When "formula" is specified, all terms used
for training the model are referenced in the IntMatrix field of the
obj class object as a matrix containing the column indexes for each
term including both the predictors and the interactions used. | |
"interactions" | a logical matrix, a positive integer
scalar, or the string "all" for defining the interactions between
predictor variables. When given a logical matrix, it must have the same
number of columns as X and each row corresponds to a different
interaction term combining the predictors indexed as true. Each
interaction term is appended as a column vector after the available predictor
column in X. When "all" is defined, then all possible
combinations of interactions are appended in X before training. At the
moment, parsing a positive integer has the same effect as the "all"
option. When "interactions" is specified, only the interaction terms
appended to X are referenced in the IntMatrix field of the
obj class object. | |
"knots" | a scalar or a row vector with the same
columns as X. It defines the knots for fitting a polynomial when
training the GAM. As a scalar, it is expanded to a row vector. The default
value is 5, hence expanded to ones (1, columns (X)) * 5. You can
parse a row vector with different number of knots for each predictor
variable to be fitted with, although not recommended. | |
"order" | a scalar or a row vector with the same
columns as X. It defines the order of the polynomial when training the
GAM. As a scalar, it is expanded to a row vector. The default values is 3,
hence expanded to ones (1, columns (X)) * 3. You can parse a row
vector with different number of polynomial order for each predictor variable
to be fitted with, although not recommended. | |
"dof" | a scalar or a row vector with the same columns
as X. It defines the degrees of freedom for fitting a polynomial when
training the GAM. As a scalar, it is expanded to a row vector. The default
value is 8, hence expanded to ones (1, columns (X)) * 8. You can
parse a row vector with different degrees of freedom for each predictor
variable to be fitted with, although not recommended. | |
"tol" | a positive scalar to set the tolerance for
convergence during training. By default, it is set to 1e-3. |
You can parse either a "formula" or an "interactions"
optional parameter. Parsing both parameters will result an error.
Accordingly, you can only pass up to two parameters among "knots",
"order", and "dof" to define the required polynomial for
training the GAM model.
See also: fitrgam, regress, regress_gp
Source Code: RegressionGAM
Properties
RegressionGAM.X is not documented.
RegressionGAM.Y is not documented.
RegressionGAM.BaseModel is not documented.
RegressionGAM.ModelwInt is not documented.
RegressionGAM.IntMatrix is not documented.
RegressionGAM.NumObservations is not documented.
RegressionGAM.RowsUsed is not documented.
RegressionGAM.NumPredictors is not documented.
RegressionGAM.PredictorNames is not documented.
RegressionGAM.ResponseName is not documented.
RegressionGAM.Formula is not documented.
RegressionGAM.Interactions is not documented.
RegressionGAM.Knots is not documented.
RegressionGAM.Order is not documented.
RegressionGAM.DoF is not documented.
RegressionGAM.Tol is not documented.
Methods
RegressionGAM: yFit = predict (obj, Xfit)
RegressionGAM: yFit = predict (…, Name, Value)
RegressionGAM: [yFit, ySD, yInt] = predict (…)
yFit = predict (obj, Xfit returns a vector of
predicted responses, yFit, for the predictor data in matrix Xfit
based on the Generalized Additive Model in obj. Xfit must have
the same number of features/variables as the training data in obj.
-
obj must be a
RegressionGAMclass object.
[yFit, ySD, yInt] = predict (obj, Xfit
also returns the standard deviations, ySD, and prediction intervals,
yInt, of the response variable yFit, evaluated at each
observation in the predictor data Xfit.
yFit = predict (…, Name, Value) returns the
aforementioned results with additional properties specified by
Name-Value pair arguments listed below.
| Name | Value | |
|---|---|---|
"alpha" | significance level of the prediction
intervals yInt, specified as scalar in range [0,1]. The default
value is 0.05, which corresponds to 95% prediction intervals. | |
"includeinteractions" | a boolean flag to include
interactions to predict new values based on Xfit. By default,
"includeinteractions" is true when the GAM model in obj
contains a obj.Formula or obj.Interactions fields. Otherwise,
is set to false. If set to true when no interactions are
present in the trained model, it will result to an error. If set to
false when using a model that includes interactions, the predictions
will be made on the basic model without any interaction terms. This way you
can make predictions from the same GAM model without having to retrain it. |
See also: fitrgam, RegressionGAM
RegressionGAM: savemodel (obj, filename)
savemodel (obj, filename) saves a RegressionGAM
object into a file defined by filename.
See also: loadmodel, fitrgam, RegressionGAM
Examples
f1 = @(x) cos (3 * x); f2 = @(x) x .^ 3; x1 = 2 * rand (50, 1) - 1; x2 = 2 * rand (50, 1) - 1; y = f1(x1) + f2(x2); y = y + y .* 0.2 .* rand (50,1); X = [x1, x2]; a = fitrgam (X, y, "tol", 1e-3) |
a =
1x1 RegressionGAM object with properties:
X: [0.2789, -0.2596; -0.9500, -0.5810; -0.4499, -0.4660; -0.5536, 0.8733; 0.4729, 0.2961; ...]
Y: [0.6540; -1.3202; 0.1340; 0.6381; 0.1868; 0.5631; -1.0115; -0.7519; 0.6405; -1.1469; ...]
BaseModel: [1x1 struct]
ModelwInt: []
IntMatrix: []
NumObservations: 50
RowsUsed: [1; 1; 1; 1; 1; 1; 1; 1; 1; 1; ...]
NumPredictors: 2
PredictorNames: [1x2 cell]
ResponseName: "Y"
Formula: []
Interactions: []
Knots: [5, 5]
Order: [3, 3]
DoF: [8, 8]
Tol: 1.0000e-03
|
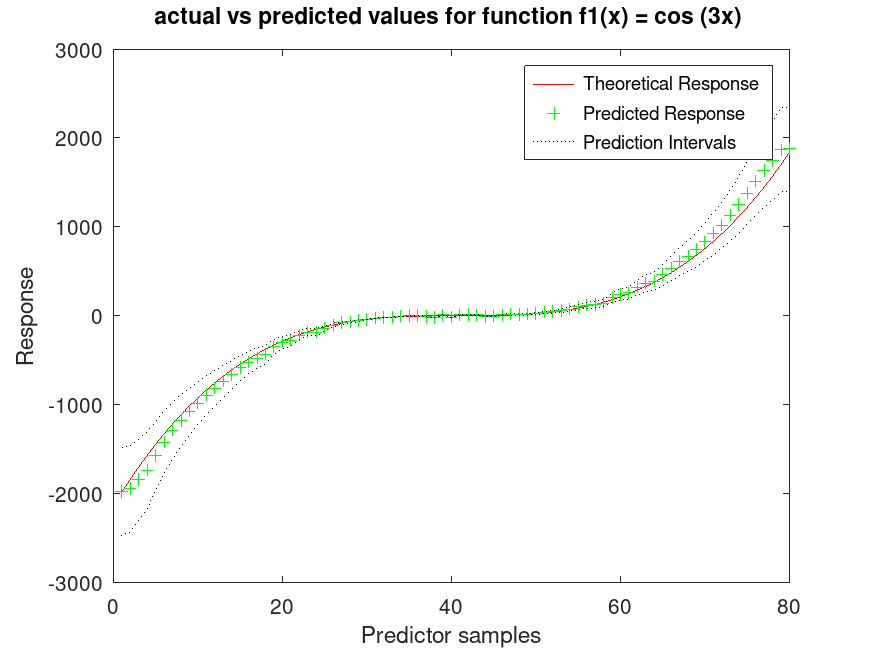
f1 = @(x) cos (3 * x);
f2 = @(x) x .^ 3;
## Generate 80 samples for f1 and f2
x = [-4*pi:0.1*pi:4*pi-0.1*pi]';
X1 = f1 (x);
X2 = f2 (x);
## Create a synthetic response by adding noise
rand ("seed", 3);
Ytrue = X1 + X2;
Y = Ytrue + Ytrue .* 0.2 .* rand (80,1);
## Assemble predictor data
X = [X1, X2];
## Train the GAM and test on the same data
a = fitrgam (X, Y, "order", [5, 5]);
[ypred, ySDsd, yInt] = predict (a, X);
## Plot the results
figure
[sortedY, indY] = sort (Ytrue);
plot (sortedY, "r-");
xlim ([0, 80]);
hold on
plot (ypred(indY), "g+")
plot (yInt(indY,1), "k:")
plot (yInt(indY,2), "k:")
xlabel ("Predictor samples");
ylabel ("Response");
title ("actual vs predicted values for function f1(x) = cos (3x) ");
legend ({"Theoretical Response", "Predicted Response", "Prediction Intervals"});
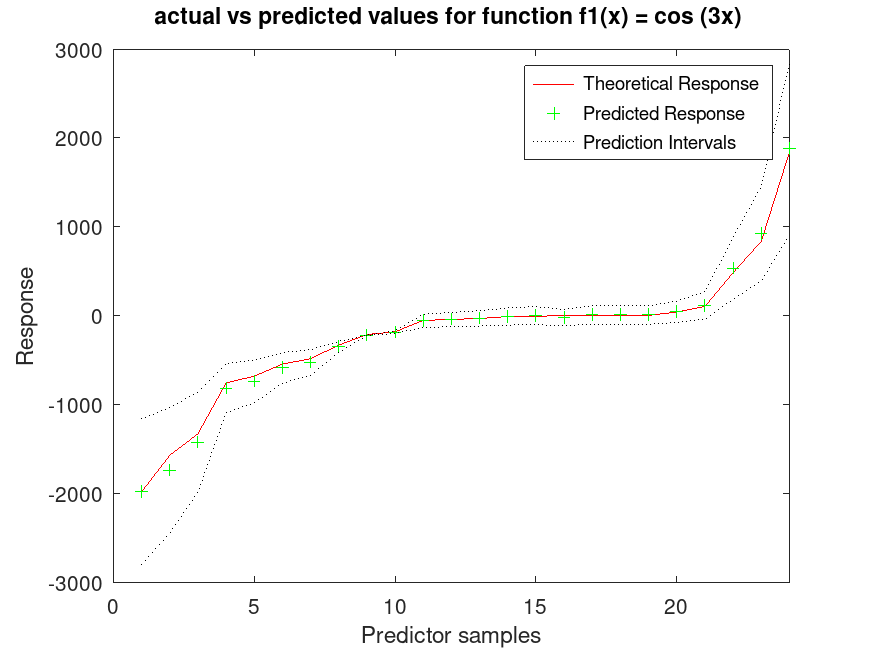
## Use 30% Holdout partitioning for training and testing data
C = cvpartition (80, "HoldOut", 0.3);
[ypred, ySDsd, yInt] = predict (a, X(test(C),:));
## Plot the results
figure
[sortedY, indY] = sort (Ytrue(test(C)));
plot (sortedY, 'r-');
xlim ([0, sum(test(C))]);
hold on
plot (ypred(indY), "g+")
plot (yInt(indY,1),'k:')
plot (yInt(indY,2),'k:')
xlabel ("Predictor samples");
ylabel ("Response");
title ("actual vs predicted values for function f1(x) = cos (3x) ");
legend ({"Theoretical Response", "Predicted Response", "Prediction Intervals"}); |