Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: fitcsvm
statistics: Mdl = fitcsvm (X, Y)
statistics: Mdl = fitcsvm (…, name, value)
Fit a Support Vector Machine classification model.
Mdl = fitcsvm (X, Y) returns a Support Vector
Machine classification model, Mdl, with X being the predictor
data, and Y the class labels of observations in X.
-
Xmust be a numeric matrix of predictor data where rows correspond to observations and columns correspond to features or variables. -
Yis matrix or cell matrix containing the class labels of corresponding predictor data in X. Y can be numerical, logical, char array or cell array of character vectors. Y must have same number of rows as X.
Mdl = fitcsvm (…, name, value) returns a
Support Vector Machine model with additional options specified by
Name-Value pair arguments listed below.
Model Parameters
| Name | Value | |
|---|---|---|
"Standardize" | A boolean flag indicating whether the data in X should be standardized prior to training. | |
"PredictorNames" | A cell array of character vectors specifying the predictor variable names. The variable names are assumed to be in the same order as they appear in the training data X. | |
"ResponseName" | A character vector specifying the name of the response variable. | |
"ClassNames" | Names of the classes in the class
labels, Y, used for fitting the kNN model. ClassNames are of
the same type as the class labels in Y. | |
"SVMtype" | Specifies the type of SVM used for training
the ClassificationSVM model. By default, the type of SVM is defined
by setting other parameters and/or by the data itself. Setting the
"SVMtype" parameter overrides the default behavior and it accepts the
following options: |
| Value | Description | |
|---|---|---|
"C_SVC" | It is the standard SVM formulation for
classification tasks. It aims to find the optimal hyperplane that separates
different classes by maximizing the margin between them while allowing some
misclassifications. The parameter "C" controls the trade-off between
maximizing the margin and minimizing the classification error. It is the
default type, unless otherwise specified. | |
"nu_SVC" | It is a variation of the standard SVM that
introduces a parameter (nu) as an upper bound on the fraction of
margin errors and a lower bound on the fraction of support vectors. This
formulation provides more control over the number of support vectors and the
margin errors, making it useful for specific classification scenarios. It is
the default type, when the "OutlierFraction" parameter is set. | |
"one_class_SVM" | It is used for anomaly detection and
novelty detection tasks. It aims to separate the data points of a single
class from the origin in a high-dimensional feature space. This method is
particularly useful for identifying outliers or unusual patterns in the data.
It is the default type, when the "Nu" parameter is set or when there
is a single class in Y. When "one_class_SVM" is set by the
"SVMtype" pair argument, Y has no effect and any classes are
ignored. |
| Name | Value | |
|---|---|---|
"OutlierFraction" | The expected proportion of outliers
in the training data, specified as a scalar value in the range .
When specified, the type of SVM model is switched to "nu_SVC" and
"OutlierFraction" defines the (nu) parameter. | |
"KernelFunction" | A character vector specifying the
method for computing elements of the Gram matrix. The available kernel
functions are 'gaussian' or 'rbf', 'linear',
'polynomial', and 'sigmoid'. For one-class learning, the
default Kernel function is 'rbf'. For two-class learning the default
is 'linear'. | |
"PolynomialOrder" | A positive integer that specifies
the order of polynomial in kernel function. The default value is 3. Unless
the "KernelFunction" is set to 'polynomial', this parameter
is ignored. | |
"KernelScale" | A positive scalar that specifies a
scaling factor for the (gamma) parameter, which can be seen as the
inverse of the radius of influence of samples selected by the model as
support vectors. The (gamma) parameter is computed as
. The default value
for "KernelScale" is 1. | |
"KernelOffset" | A nonnegative scalar that specifies
the in kernel function. For the polynomial kernel, it influences
the polynomial’s shift, and for the sigmoid kernel, it affects the hyperbolic
tangent’s shift. The default value for "KernelOffset" is 0. | |
"BoxConstraint" | A positive scalar that specifies the
upper bound of the Lagrange multipliers, i.e. the parameter C, which is used
for training "C_SVC" and "one_class_SVM" type of models. It
determines the trade-off between maximizing the margin and minimizing the
classification error. The default value for "BoxConstraint" is 1. | |
"Nu" | A positive scalar, in the range
that specifies the parameter (nu) for training "nu_SVC" and
"one_class_SVM" type of models. Unless overridden by setting the
"SVMtype" parameter, setting the "Nu" parameter always forces
the training model type to "one_class_SVM", in which case, the number
of classes in Y is ignored. The default value for "Nu" is 1. | |
"CacheSize" | A positive scalar that specifies the memory requirements (in MB) for storing the Gram matrix. The default is 1000. | |
"Tolerance" | A nonnegative scalar that specifies the tolerance of termination criterion. The default value is 1e-6. | |
"Shrinking" | Specifies whether to use shrinking heuristics. It accepts either 0 or 1. The default value is 1. |
Cross Validation Options
| Name | Value | |
|---|---|---|
"Crossval" | Cross-validation flag specified as
'on' or 'off'. If 'on' is specified, a 10-fold
cross validation is performed and a ClassificationPartitionedModel is
returned in Mdl. To override this cross-validation setting, use only
one of the following Name-Value pair arguments. | |
"CVPartition" | A cvpartition object that
specifies the type of cross-validation and the indexing for the training and
validation sets. A ClassificationPartitionedModel is returned in
Mdl and the trained model is stored in the Trained property. | |
"Holdout" | Fraction of the data used for holdout
validation, specified as a scalar value in the range . When
specified, a randomly selected percentage is reserved as validation data and
the remaining set is used for training. The trained model is stored in the
Trained property of the ClassificationPartitionedModel returned
in Mdl. "Holdout" partitioning attempts to ensure that each
partition represents the classes proportionately. | |
"KFold" | Number of folds to use in the cross-validated
model, specified as a positive integer value greater than 1. When specified,
then the data is randomly partitioned in sets and for each set, the
set is reserved as validation data while the remaining sets are
used for training. The trained models are stored in the Trained
property of the ClassificationPartitionedModel returned in Mdl.
"KFold" partitioning attempts to ensure that each partition
represents the classes proportionately. | |
"Leaveout" | Leave-one-out cross-validation flag
specified as 'on' or 'off'. If 'on' is specified,
then for each of the observations (where is the number of
observations, excluding missing observations, specified in the
NumObservations property of the model), one observation is reserved as
validation data while the remaining observations are used for training. The
trained models are stored in the Trained property of the
ClassificationPartitionedModel returned in Mdl. |
See also: ClassificationSVM, ClassificationPartitionedModel, svmtrain, svmpredict
Source Code: fitcsvm
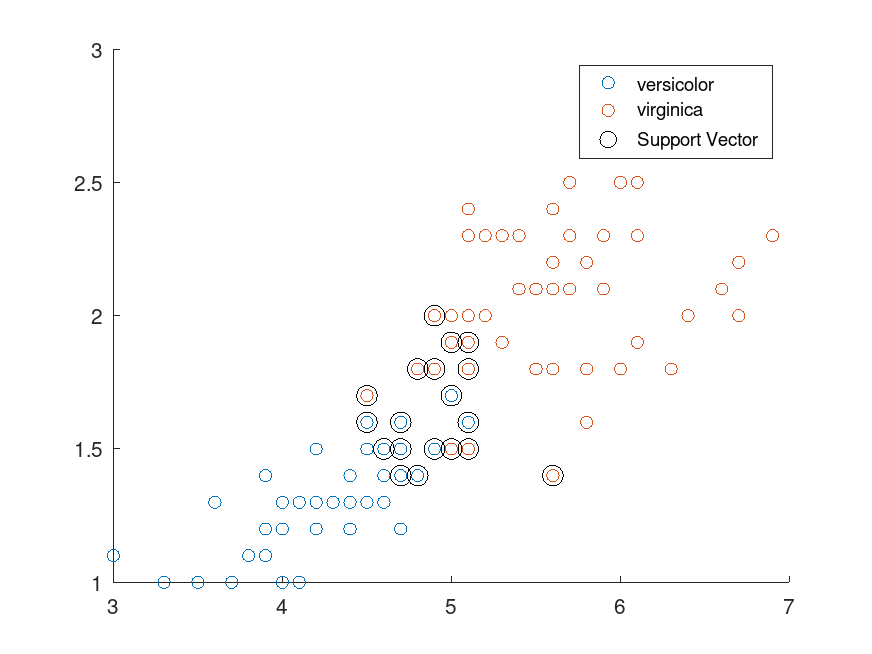
Example: 1
## Use a subset of Fisher's iris data set
load fisheriris
inds = ! strcmp (species, 'setosa');
X = meas(inds, [3,4]);
Y = species(inds);
## Train a linear SVM classifier
SVMModel = fitcsvm (X, Y)
## Plot a scatter diagram of the data and circle the support vectors.
sv = SVMModel.SupportVectors;
figure
gscatter (X(:,1), X(:,2), Y)
hold on
plot (sv(:,1), sv(:,2), 'ko', 'MarkerSize', 10)
legend ('versicolor', 'virginica', 'Support Vector')
hold off
SVMModel =
ClassificationSVM
ResponseName: 'Y'
ClassNames: {'versicolor' 'virginica'}
ScoreTransform: 'none'
NumObservations: 100
NumPredictors: 2
Alpha: [24x1 double]
Bias: -14.414868
KernelParameters: [1x1 struct]
ans =
-82.359 -81.820
|