Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: fitcknn
statistics: Mdl = fitcknn (X, Y)
statistics: Mdl = fitcknn (…, name, value)
Fit a k-Nearest Neighbor classification model.
Mdl = fitcknn (X, Y) returns a k-Nearest Neighbor
classification model, Mdl, with X being the predictor data, and
Y the class labels of observations in X.
-
Xmust be a numeric matrix of predictor data where rows correspond to observations and columns correspond to features or variables. -
Yis matrix or cell matrix containing the class labels of corresponding predictor data in X. Y can be numerical, logical, char array or cell array of character vectors. Y must have same number of rows as X.
Mdl = fitcknn (…, name, value) returns a
k-Nearest Neighbor classification model with additional options specified by
Name-Value pair arguments listed below.
Model Parameters
| Name | Value | |
|---|---|---|
"Standardize" | A boolean flag indicating whether the data in X should be standardized prior to training. | |
"PredictorNames" | A cell array of character vectors specifying the predictor variable names. The variable names are assumed to be in the same order as they appear in the training data X. | |
"ResponseName" | A character vector specifying the name of the response variable. | |
"ClassNames" | Names of the classes in the class
labels, Y, used for fitting the kNN model. ClassNames are of
the same type as the class labels in Y. | |
"Prior" | A numeric vector specifying the prior
probabilities for each class. The order of the elements in Prior
corresponds to the order of the classes in ClassNames. | |
"Cost" | A numeric matrix containing
misclassification cost for the corresponding instances in X where
is the number of unique categories in Y. If an instance is
correctly classified into its category the cost is calculated to be 1,
otherwise 0. cost matrix can be altered use Mdl.cost = somecost.
default value cost = ones(rows(X),numel(unique(Y))). | |
"ScoreTransform" | A character vector defining one of
the following functions or a user defined function handle, which is used
for transforming the prediction scores returned by the predict and
resubPredict methods. Default value is 'none'. |
| Value | Description | |
|---|---|---|
"doublelogit" | ||
"invlogit" | ||
"ismax" | Sets the score for the class with the largest score to 1, and sets the scores for all other classes to 0 | |
"logit" | ||
"none" | (no transformation) | |
"identity" | (no transformation) | |
"sign" | ||
"symmetric" | ||
"symmetricismax" | Sets the score for the class with the largest score to 1, and sets the scores for all other classes to -1 | |
"symmetriclogit" |
| Name | Value | |
|---|---|---|
"BreakTies" | Tie-breaking algorithm used by predict when multiple classes have the same smallest cost. By default, ties occur when multiple classes have the same number of nearest points among the nearest neighbors. The available options are specified by the following character arrays: |
| Value | Description | |
|---|---|---|
"smallest" | This is the default and it favors the class with the smallest index among the tied groups, i.e. the one that appears first in the training labelled data. | |
"nearest" | This favors the class with the nearest neighbor among the tied groups, i.e. the class with the closest member point according to the distance metric used. | |
"random" | This randomly picks one class among the tied groups. |
| Name | Value | |
|---|---|---|
"BucketSize" | The maximum number of data points in the
leaf node of the Kd-tree and it must be a positive integer. By default, it
is 50. This argument is meaningful only when the selected search method is
"kdtree". | |
"NumNeighbors" | A positive integer value specifying the number of nearest neighbors to be found in the kNN search. By default, it is 1. | |
"Exponent" | A positive scalar (usually an integer)
specifying the Minkowski distance exponent. This argument is only valid when
the selected distance metric is "minkowski". By default it is 2. | |
"Scale" | A nonnegative numeric vector specifying the
scale parameters for the standardized Euclidean distance. The vector length
must be equal to the number of columns in X. This argument is only
valid when the selected distance metric is "seuclidean", in which
case each coordinate of X is scaled by the corresponding element of
"scale", as is each query point in Y. By default, the scale
parameter is the standard deviation of each coordinate in X. If a
variable in X is constant, i.e. zero variance, this value is forced
to 1 to avoid division by zero. This is the equivalent of this variable not
being standardized. | |
"Cov" | A square matrix with the same number of columns
as X specifying the covariance matrix for computing the mahalanobis
distance. This must be a positive definite matrix matching. This argument
is only valid when the selected distance metric is "mahalanobis". | |
"Distance" | is the distance metric used by
knnsearch as specified below: |
| Value | Description | |
|---|---|---|
"euclidean" | Euclidean distance. | |
"seuclidean" | standardized Euclidean distance. Each
coordinate difference between the rows in X and the query matrix
Y is scaled by dividing by the corresponding element of the standard
deviation computed from X. To specify a different scaling, use the
"Scale" name-value argument. | |
"cityblock" | City block distance. | |
"chebychev" | Chebychev distance (maximum coordinate difference). | |
"minkowski" | Minkowski distance. The default exponent
is 2. To specify a different exponent, use the "P" name-value
argument. | |
"mahalanobis" | Mahalanobis distance, computed using a
positive definite covariance matrix. To change the value of the covariance
matrix, use the "Cov" name-value argument. | |
"cosine" | Cosine distance. | |
"correlation" | One minus the sample linear correlation between observations (treated as sequences of values). | |
"spearman" | One minus the sample Spearman’s rank correlation between observations (treated as sequences of values). | |
"hamming" | Hamming distance, which is the percentage of coordinates that differ. | |
"jaccard" | One minus the Jaccard coefficient, which is the percentage of nonzero coordinates that differ. | |
| @distfun | Custom distance function handle. A distance
function of the form function D2 = distfun (XI, YI),
where XI is a vector containing a single observation in
-dimensional space, YI is an matrix containing an
arbitrary number of observations in the same -dimensional space, and
D2 is an vector of distances, where (D2k) is
the distance between observations XI and (YIk,:). |
| Name | Value | |
|---|---|---|
"DistanceWeight" | A distance weighting function,
specified either as a function handle, which accepts a matrix of nonnegative
distances and returns a matrix the same size containing nonnegative distance
weights, or one of the following values: "equal", which corresponds
to no weighting; "inverse", which corresponds to a weight equal to
; "squaredinverse", which corresponds to a weight
equal to . | |
"IncludeTies" | A boolean flag to indicate if the
returned values should contain the indices that have same distance as the
neighbor. When false, knnsearch chooses the
observation with the smallest index among the observations that have the same
distance from a query point. When true, knnsearch includes
all nearest neighbors whose distances are equal to the smallest
distance in the output arguments. To specify , use the "K"
name-value pair argument. | |
"NSMethod" | is the nearest neighbor search method used
by knnsearch as specified below. |
| Value | Description | |
|---|---|---|
"kdtree" | Creates and uses a Kd-tree to find nearest
neighbors. "kdtree" is the default value when the number of columns
in X is less than or equal to 10, X is not sparse, and the
distance metric is "euclidean", "cityblock",
"manhattan", "chebychev", or "minkowski". Otherwise,
the default value is "exhaustive". This argument is only valid when
the distance metric is one of the four aforementioned metrics. | |
"exhaustive" | Uses the exhaustive search algorithm by computing the distance values from all the points in X to each point in Y. |
Cross Validation Options
| Name | Value | |
|---|---|---|
"Crossval" | Cross-validation flag specified as
'on' or 'off'. If 'on' is specified, a 10-fold
cross validation is performed and a ClassificationPartitionedModel is
returned in Mdl. To override this cross-validation setting, use only
one of the following Name-Value pair arguments. | |
"CVPartition" | A cvpartition object that
specifies the type of cross-validation and the indexing for the training and
validation sets. A ClassificationPartitionedModel is returned in
Mdl and the trained model is stored in the Trained property. | |
"Holdout" | Fraction of the data used for holdout
validation, specified as a scalar value in the range . When
specified, a randomly selected percentage is reserved as validation data and
the remaining set is used for training. The trained model is stored in the
Trained property of the ClassificationPartitionedModel returned
in Mdl. "Holdout" partitioning attempts to ensure that each
partition represents the classes proportionately. | |
"KFold" | Number of folds to use in the cross-validated
model, specified as a positive integer value greater than 1. When specified,
then the data is randomly partitioned in sets and for each set, the
set is reserved as validation data while the remaining sets are
used for training. The trained models are stored in the Trained
property of the ClassificationPartitionedModel returned in Mdl.
"KFold" partitioning attempts to ensure that each partition
represents the classes proportionately. | |
"Leaveout" | Leave-one-out cross-validation flag
specified as 'on' or 'off'. If 'on' is specified,
then for each of the observations (where is the number of
observations, excluding missing observations, specified in the
NumObservations property of the model), one observation is reserved as
validation data while the remaining observations are used for training. The
trained models are stored in the Trained property of the
ClassificationPartitionedModel returned in Mdl. |
See also: ClassificationKNN, ClassificationPartitionedModel, knnsearch, rangesearch, pdist2
Source Code: fitcknn
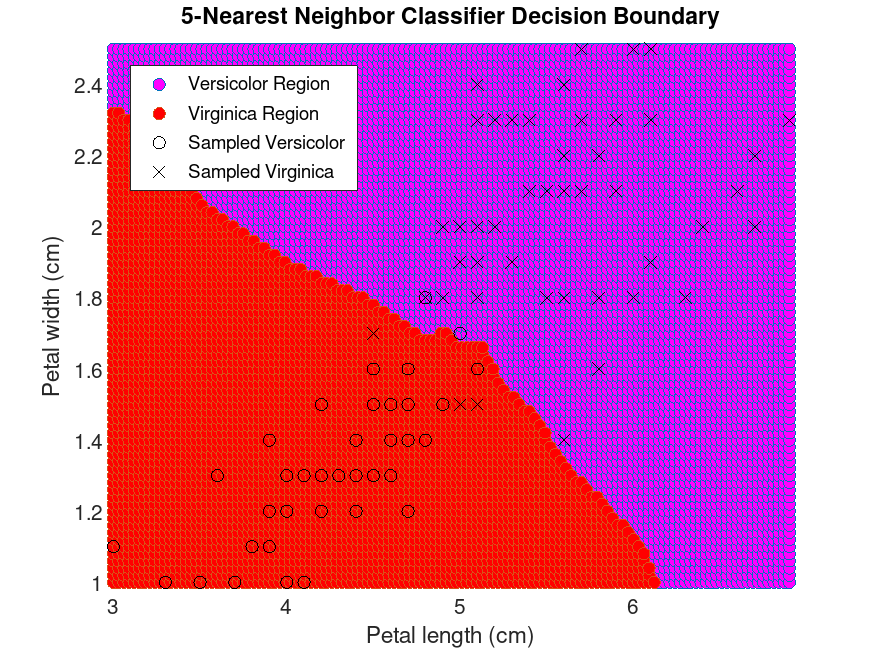
Example: 1
## Train a k-nearest neighbor classifier for k = 10
## and plot the decision boundaries.
load fisheriris
idx = ! strcmp (species, "setosa");
X = meas(idx,3:4);
Y = cast (strcmpi (species(idx), "virginica"), "double");
obj = fitcknn (X, Y, "Standardize", 1, "NumNeighbors", 10, "NSMethod", "exhaustive")
x1 = [min(X(:,1)):0.03:max(X(:,1))];
x2 = [min(X(:,2)):0.02:max(X(:,2))];
[x1G, x2G] = meshgrid (x1, x2);
XGrid = [x1G(:), x2G(:)];
pred = predict (obj, XGrid);
gidx = logical (pred);
figure
scatter (XGrid(gidx,1), XGrid(gidx,2), "markerfacecolor", "magenta");
hold on
scatter (XGrid(!gidx,1), XGrid(!gidx,2), "markerfacecolor", "red");
plot (X(Y == 0, 1), X(Y == 0, 2), "ko", X(Y == 1, 1), X(Y == 1, 2), "kx");
xlabel ("Petal length (cm)");
ylabel ("Petal width (cm)");
title ("5-Nearest Neighbor Classifier Decision Boundary");
legend ({"Versicolor Region", "Virginica Region", ...
"Sampled Versicolor", "Sampled Virginica"}, ...
"location", "northwest")
axis tight
hold off
obj =
ClassificationKNN
ResponseName: 'Y'
ClassNames: [0 1]
ScoreTransform: 'none'
NumObservations: 100
NumPredictors: 2
Distance: 'euclidean'
NSMethod: 'exhaustive'
NumNeighbors: 10
|