Categories &
Functions List
- BetaDistribution
- BinomialDistribution
- BirnbaumSaundersDistribution
- BurrDistribution
- ExponentialDistribution
- ExtremeValueDistribution
- GammaDistribution
- GeneralizedExtremeValueDistribution
- GeneralizedParetoDistribution
- HalfNormalDistribution
- InverseGaussianDistribution
- LogisticDistribution
- LoglogisticDistribution
- LognormalDistribution
- LoguniformDistribution
- MultinomialDistribution
- NakagamiDistribution
- NegativeBinomialDistribution
- NormalDistribution
- PiecewiseLinearDistribution
- PoissonDistribution
- RayleighDistribution
- RicianDistribution
- tLocationScaleDistribution
- TriangularDistribution
- UniformDistribution
- WeibullDistribution
- betafit
- betalike
- binofit
- binolike
- bisafit
- bisalike
- burrfit
- burrlike
- evfit
- evlike
- expfit
- explike
- gamfit
- gamlike
- geofit
- gevfit_lmom
- gevfit
- gevlike
- gpfit
- gplike
- gumbelfit
- gumbellike
- hnfit
- hnlike
- invgfit
- invglike
- logifit
- logilike
- loglfit
- logllike
- lognfit
- lognlike
- nakafit
- nakalike
- nbinfit
- nbinlike
- normfit
- normlike
- poissfit
- poisslike
- raylfit
- rayllike
- ricefit
- ricelike
- tlsfit
- tlslike
- unidfit
- unifit
- wblfit
- wbllike
- betacdf
- betainv
- betapdf
- betarnd
- binocdf
- binoinv
- binopdf
- binornd
- bisacdf
- bisainv
- bisapdf
- bisarnd
- burrcdf
- burrinv
- burrpdf
- burrrnd
- bvncdf
- bvtcdf
- cauchycdf
- cauchyinv
- cauchypdf
- cauchyrnd
- chi2cdf
- chi2inv
- chi2pdf
- chi2rnd
- copulacdf
- copulapdf
- copularnd
- evcdf
- evinv
- evpdf
- evrnd
- expcdf
- expinv
- exppdf
- exprnd
- fcdf
- finv
- fpdf
- frnd
- gamcdf
- gaminv
- gampdf
- gamrnd
- geocdf
- geoinv
- geopdf
- geornd
- gevcdf
- gevinv
- gevpdf
- gevrnd
- gpcdf
- gpinv
- gppdf
- gprnd
- gumbelcdf
- gumbelinv
- gumbelpdf
- gumbelrnd
- hncdf
- hninv
- hnpdf
- hnrnd
- hygecdf
- hygeinv
- hygepdf
- hygernd
- invgcdf
- invginv
- invgpdf
- invgrnd
- iwishpdf
- iwishrnd
- jsucdf
- jsupdf
- laplacecdf
- laplaceinv
- laplacepdf
- laplacernd
- logicdf
- logiinv
- logipdf
- logirnd
- loglcdf
- loglinv
- loglpdf
- loglrnd
- logncdf
- logninv
- lognpdf
- lognrnd
- mnpdf
- mnrnd
- mvncdf
- mvnpdf
- mvnrnd
- mvtcdf
- mvtpdf
- mvtrnd
- mvtcdfqmc
- nakacdf
- nakainv
- nakapdf
- nakarnd
- nbincdf
- nbininv
- nbinpdf
- nbinrnd
- ncfcdf
- ncfinv
- ncfpdf
- ncfrnd
- nctcdf
- nctinv
- nctpdf
- nctrnd
- ncx2cdf
- ncx2inv
- ncx2pdf
- ncx2rnd
- normcdf
- norminv
- normpdf
- normrnd
- plcdf
- plinv
- plpdf
- plrnd
- poisscdf
- poissinv
- poisspdf
- poissrnd
- raylcdf
- raylinv
- raylpdf
- raylrnd
- ricecdf
- riceinv
- ricepdf
- ricernd
- tcdf
- tinv
- tpdf
- trnd
- tlscdf
- tlsinv
- tlspdf
- tlsrnd
- tricdf
- triinv
- tripdf
- trirnd
- unidcdf
- unidinv
- unidpdf
- unidrnd
- unifcdf
- unifinv
- unifpdf
- unifrnd
- vmcdf
- vminv
- vmpdf
- vmrnd
- wblcdf
- wblinv
- wblpdf
- wblrnd
- wienrnd
- wishpdf
- wishrnd
- adtest
- anova1
- anova2
- anovan
- bartlett_test
- barttest
- binotest
- chi2gof
- chi2test
- correlation_test
- fishertest
- friedman
- hotelling_t2test
- hotelling_t2test2
- kruskalwallis
- kstest
- kstest2
- levene_test
- manova1
- mcnemar_test
- multcompare
- ranksum
- regression_ftest
- regression_ttest
- runstest
- sampsizepwr
- signrank
- signtest
- tiedrank
- ttest
- ttest2
- vartest
- vartest2
- vartestn
- ztest
- ztest2
Function Reference: rangesearch
statistics: idx = rangesearch (X, Y, r)
statistics: [idx, D] = rangesearch (X, Y, r)
statistics: […] = rangesearch (…, name, value)
Find all neighbors within specified distance from input data.
idx = rangesearch (X, Y, r) returns all the
points in X that are within distance r from the points in Y.
X must be an numeric matrix of input data, where rows
correspond to observations and columns correspond to features or variables.
Y is an numeric matrix with query points, which must have
the same numbers of column as X. r must be a nonnegative scalar
value. idx is an cell array, where is the number
of observations in Y. The vector Idx{j} contains the
indices of observations (rows) in X whose distances to
Y(j,:) are not greater than r.
[idx, D] = rangesearch (X, Y, r) also
returns the distances, D, which correspond to the points in X
that are within distance r from the points in Y. D is an
cell array, where is the number of observations in
Y. The vector D{j} contains the distances of
observations (rows) in X whose distances to Y(j,:) are
not greater than r.
Additional parameters can be specified by Name-Value pair arguments.
| Name | Value | |
|---|---|---|
"P" | is the Minkowski distance exponent and it must be
a positive scalar. This argument is only valid when the selected distance
metric is "minkowski". By default it is 2. | |
"Scale" | is the scale parameter for the standardized
Euclidean distance and it must be a nonnegative numeric vector of equal
length to the number of columns in X. This argument is only valid when
the selected distance metric is "seuclidean", in which case each
coordinate of X is scaled by the corresponding element of
"scale", as is each query point in Y. By default, the scale
parameter is the standard deviation of each coordinate in X. | |
"Cov" | is the covariance matrix for computing the
mahalanobis distance and it must be a positive definite matrix matching the
the number of columns in X. This argument is only valid when the
selected distance metric is "mahalanobis". | |
"BucketSize" | is the maximum number of data points in
the leaf node of the Kd-tree and it must be a positive integer. This
argument is only valid when the selected search method is "kdtree". | |
"SortIndices" | is a boolean flag to sort the returned
indices in ascending order by distance and it is true by default.
When the selected search method is "exhaustive" or the
"IncludeTies" flag is true, rangesearch always sorts the
returned indices. | |
"Distance" | is the distance metric used by
rangesearch as specified below: |
"euclidean" | Euclidean distance. | |
"seuclidean" | standardized Euclidean distance. Each
coordinate difference between the rows in X and the query matrix
Y is scaled by dividing by the corresponding element of the standard
deviation computed from X. To specify a different scaling, use the
"Scale" name-value argument. | |
"cityblock" | City block distance. | |
"chebychev" | Chebychev distance (maximum coordinate difference). | |
"minkowski" | Minkowski distance. The default exponent
is 2. To specify a different exponent, use the "P" name-value
argument. | |
"mahalanobis" | Mahalanobis distance, computed using a
positive definite covariance matrix. To change the value of the covariance
matrix, use the "Cov" name-value argument. | |
"cosine" | Cosine distance. | |
"correlation" | One minus the sample linear correlation between observations (treated as sequences of values). | |
"spearman" | One minus the sample Spearman’s rank correlation between observations (treated as sequences of values). | |
"hamming" | Hamming distance, which is the percentage of coordinates that differ. | |
"jaccard" | One minus the Jaccard coefficient, which is the percentage of nonzero coordinates that differ. | |
| @distfun | Custom distance function handle. A distance
function of the form function D2 = distfun (XI, YI),
where XI is a vector containing a single observation in
-dimensional space, YI is an matrix containing an
arbitrary number of observations in the same -dimensional space, and
D2 is an vector of distances, where (D2k) is
the distance between observations XI and (YIk,:). |
"NSMethod" | is the nearest neighbor search method used
by rangesearch as specified below. |
"kdtree" | Creates and uses a Kd-tree to find nearest
neighbors. "kdtree" is the default value when the number of columns
in X is less than or equal to 10, X is not sparse, and the
distance metric is "euclidean", "cityblock",
"manhattan", "chebychev", or "minkowski". Otherwise,
the default value is "exhaustive". This argument is only valid when
the distance metric is one of the four aforementioned metrics. | |
"exhaustive" | Uses the exhaustive search algorithm by computing the distance values from all the points in X to each point in Y. |
Source Code: rangesearch
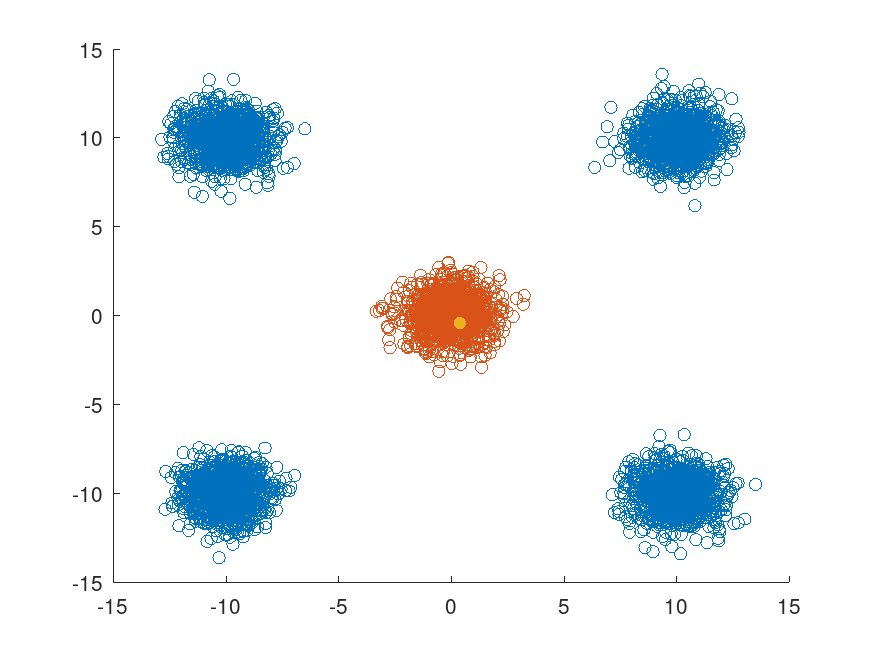
Example: 1
## Generate 1000 random 2D points from each of five distinct multivariate
## normal distributions that form five separate classes
N = 1000;
d = 10;
randn ("seed", 5);
X1 = mvnrnd (d * [0, 0], eye (2), 1000);
randn ("seed", 6);
X2 = mvnrnd (d * [1, 1], eye (2), 1000);
randn ("seed", 7);
X3 = mvnrnd (d * [-1, -1], eye (2), 1000);
randn ("seed", 8);
X4 = mvnrnd (d * [1, -1], eye (2), 1000);
randn ("seed", 8);
X5 = mvnrnd (d * [-1, 1], eye (2), 1000);
X = [X1; X2; X3; X4; X5];
## For each point in X, find the points in X that are within a radius d
## away from the points in X.
Idx = rangesearch (X, X, d, "NSMethod", "exhaustive");
## Select the first point in X (corresponding to the first class) and find
## its nearest neighbors within the radius d. Display these points in
## one color and the remaining points in a different color.
x = X(1,:);
nearestPoints = X (Idx{1},:);
nonNearestIdx = true (size (X, 1), 1);
nonNearestIdx(Idx{1}) = false;
scatter (X(nonNearestIdx,1), X(nonNearestIdx,2))
hold on
scatter (nearestPoints(:,1),nearestPoints(:,2))
scatter (x(1), x(2), "black", "filled")
hold off
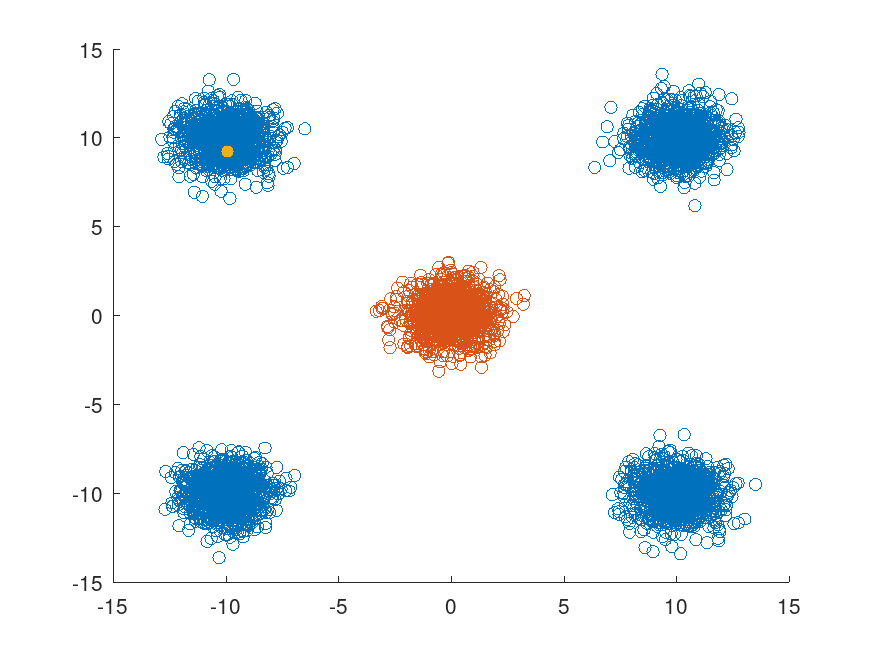
## Select the last point in X (corresponding to the fifth class) and find
## its nearest neighbors within the radius d. Display these points in
## one color and the remaining points in a different color.
x = X(end,:);
nearestPoints = X (Idx{1},:);
nonNearestIdx = true (size (X, 1), 1);
nonNearestIdx(Idx{1}) = false;
figure
scatter (X(nonNearestIdx,1), X(nonNearestIdx,2))
hold on
scatter (nearestPoints(:,1),nearestPoints(:,2))
scatter (x(1), x(2), "black", "filled")
hold off
|